1R9Y
 
 | | Bacterial cytosine deaminase D314A mutant. | | Descriptor: | Cytosine deaminase, FE (III) ION, GLYCEROL, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1RB7
 
 | |
1R9X
 
 | | Bacterial cytosine deaminase D314G mutant. | | Descriptor: | Cytosine deaminase, FE (III) ION, GLYCEROL, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1RC8
 
 | | T4 Polynucleotide Kinase bound to 5'-GTCAC-3' ssDNA | | Descriptor: | 5'-D(*GP*TP*CP*AP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-11-03 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
1RA0
 
 | | Bacterial cytosine deaminase D314G mutant bound to 5-fluoro-4-(S)-hydroxy-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1RAK
 
 | | Bacterial cytosine deaminase D314S mutant bound to 5-fluoro-4-(S)-hydroxyl-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1R9Z
 
 | | Bacterial cytosine deaminase D314S mutant. | | Descriptor: | Cytosine deaminase, FE (III) ION, GLYCEROL, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1RA5
 
 | | Bacterial cytosine deaminase D314A mutant bound to 5-fluoro-4-(S)-hydroxyl-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1RPZ
 
 | | T4 POLYNUCLEOTIDE KINASE BOUND TO 5'-TGCAC-3' SSDNA | | Descriptor: | 5'-D(*TP*GP*CP*AP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
1RRC
 
 | | T4 POLYNUCLEOTIDE KINASE BOUND TO 5'-GTC-3' SSDNA | | Descriptor: | 5'-D(*GP*TP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
1T9I
 
 | | I-CreI(D20N)/DNA complex | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Sussman, D, Otis, C, Boudreau, D, Turmel, M, Lemieux, C, Stephens, K, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Dependent DNA Cleavage Mechanism of the I-CreI LAGLIDADG Homing Endonuclease.
Biochemistry, 43, 2004
|
|
1T9J
 
 | | I-CreI(Q47E)/DNA complex | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', DNA endonuclease I-CreI | | Authors: | Chevalier, B, Sussman, D, Otis, C, Boudreau, D, Turmel, M, Lemieux, C, Stephens, K, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-Dependent DNA Cleavage Mechanism of the I-CreI LAGLIDADG Homing Endonuclease.
Biochemistry, 43, 2004
|
|
1U0C
 
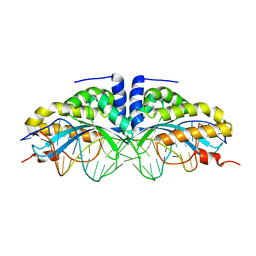 | | Y33C Mutant of Homing endonuclease I-CreI | | Descriptor: | 5'-D(*CP*GP*TP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*AP*GP*C)-3', 5'-D(*GP*CP*TP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*AP*CP*G)-3', DNA endonuclease I-CreI, ... | | Authors: | Sussman, D, Chadsey, M, Fauce, S, Engel, A, Bruett, A, Monnat, R, Stoddard, B.L, Seligman, L.M. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isolation and characterization of new homing endonuclease specificities at individual target site positions.
J.Mol.Biol., 342, 2004
|
|
3MX1
 
 | |
3MX4
 
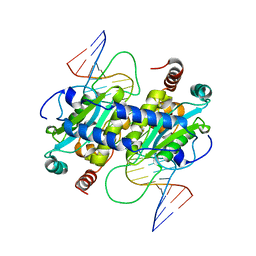 | | DNA binding and cleavage by the GIY-YIG endonuclease R.Eco29KI inactive variant E142Q | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*AP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*GP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*CP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*TP*CP*CP*CP*G)-3'), Eco29kIR | | Authors: | Mak, A.N.S, Lambert, A.R, Stoddard, B.L. | | Deposit date: | 2010-05-06 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Folding, DNA Recognition, and Function of GIY-YIG Endonucleases: Crystal Structures of R.Eco29kI.
Structure, 18, 2010
|
|
3NIC
 
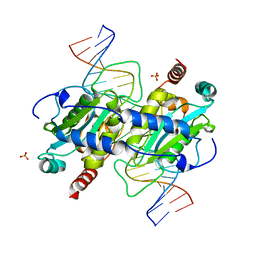 | | DNA binding and cleavage by the GIY-YIG endonuclease R.Eco29kI inactive variant Y49F | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*AP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*GP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*CP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*TP*CP*CP*CP*G)-3'), Eco29kIR, ... | | Authors: | Mak, A.N.S, Lambert, A.R, Stoddard, B.L. | | Deposit date: | 2010-06-15 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Folding, DNA Recognition, and Function of GIY-YIG Endonucleases: Crystal Structures of R.Eco29kI.
Structure, 18, 2010
|
|
3HYJ
 
 | | Crystal structure of the N-terminal LAGLIDADG domain of DUF199/WhiA | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DUF199/WhiA, ... | | Authors: | Kaiser, B.K, Clifton, M.C, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease.
Structure, 17, 2009
|
|
3HYI
 
 | | Crystal structure of full-length DUF199/WhiA from Thermatoga maritima | | Descriptor: | GLYCEROL, Protein DUF199/WhiA, SODIUM ION | | Authors: | Kaiser, B.K, Clifton, M.C, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease
Structure, 17, 2009
|
|
3I1C
 
 | |
3QQY
 
 | |
3KO2
 
 | | I-MsoI re-designed for altered DNA cleavage specificity (-7C) | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*GP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*CP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2009-11-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
3LDY
 
 | | An extraordinary mechanism of DNA perturbation exhibited by the rare-cutting HNH restriction endonuclease PacI | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*GP*GP*CP*TP*TP*A)-3'), DNA (5'-D(P*AP*TP*TP*AP*AP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Shen, B.W, Heiter, D, Chan, S.-H, Xu, S.-Y, Wilson, G, Stoddard, B.L. | | Deposit date: | 2010-01-13 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.
Structure, 18, 2010
|
|
3R7P
 
 | | The crystal structure of I-LtrI | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*TP*GP*CP*TP*CP*CP*TP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*TP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*GP*CP*AP*TP*TP*TP*G)-3'), ... | | Authors: | Mak, A.N.S, Takeuchi, R, Edgell, D.R, Stoddard, B.L. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Tapping natural reservoirs of homing endonucleases for targeted gene modification.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3R3P
 
 | |
3MIS
 
 | | I-MsoI re-designed for altered DNA cleavage specificity (-8G) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
