1POE
 
 | |
1RGD
 
 | | STRUCTURE REFINEMENT OF THE GLUCOCORTICOID RECEPTOR-DNA BINDING DOMAIN FROM NMR DATA BY RELAXATION MATRIX CALCULATIONS | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Van Tilborg, M.A.A, Bonvin, A.M.J.J, Hard, K, Davis, A, Maler, B, Boelens, R, Yamamoto, K.R, Kaptein, R. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the glucocorticoid receptor-DNA binding domain from NMR data by relaxation matrix calculations.
J.Mol.Biol., 247, 1995
|
|
3WRP
 
 | |
2OZ9
 
 | |
4DBB
 
 | | The PTB domain of Mint1 is autoinhibited by a helix in the C-terminal linker region | | Descriptor: | ACETIC ACID, Amyloid beta A4 precursor protein-binding family A member 1, CHLORIDE ION, ... | | Authors: | Tomchick, D.R, Rizo, J, Ho, A, Xu, Y. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Autoinhibition of Mint1 adaptor protein regulates amyloid precursor protein binding and processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4K6J
 
 | | Human cohesin inhibitor WapL | | Descriptor: | ACETATE ION, SULFATE ION, Wings apart-like protein homolog | | Authors: | Tomchick, D.R, Yu, H, Ouyang, Z. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6205 Å) | | Cite: | Structure of the human cohesin inhibitor Wapl.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4OA7
 
 | | Crystal structure of Tankyrase1 in complex with IWR1 | | Descriptor: | 4-[(3aR,4R,7S,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | Authors: | Zhang, X, He, H. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
4TOR
 
 | | Crystal structure of Tankyrase 1 with IWR-8 | | Descriptor: | 1-[(1-acetyl-5-bromo-1H-indol-6-yl)sulfonyl]-N-ethyl-N-(3-methylphenyl)piperidine-4-carboxamide, CHLORIDE ION, Tankyrase-1, ... | | Authors: | Chen, H, Zhang, X, Lum, L, Chen, C. | | Deposit date: | 2014-06-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
4OQQ
 
 | |
4OQP
 
 | |
2UXN
 
 | | Structural Basis of Histone Demethylation by LSD1 Revealed by Suicide Inactivation | | Descriptor: | CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Szewczuk, L.M, Gocke, C.B, Brautigam, C.A, Tomchick, D.R, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Histone Demethylation by Lsd1 Revealed by Suicide Inactivation.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2UXX
 
 | | Human LSD1 Histone Demethylase-CoREST in complex with an FAD- tranylcypromine adduct | | Descriptor: | CHLORIDE ION, FAD-trans-2-Phenylcyclopropylamine Adduct, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for the Inhibition of the Lsd1 Histone Demethylase by the Antidepressant Trans-2-Phenylcyclopropylamine.
Biochemistry, 46, 2007
|
|
3KZW
 
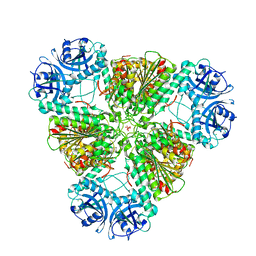 | | Crystal structure of cytosol aminopeptidase from Staphylococcus aureus COL | | Descriptor: | CHLORIDE ION, Cytosol aminopeptidase, PHOSPHATE ION, ... | | Authors: | Hattne, J, Dubrovska, I, Halavaty, A, Minasov, G, Scott, P, Shuvalova, L, Winsor, J, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: |
|
|
3KWM
 
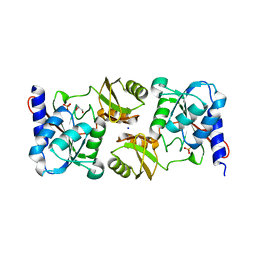 | | Crystal structure of ribose-5-isomerase A | | Descriptor: | D-Glyceraldehyde, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Orlikowska, M, Rostankowski, R, Nakka, C, Hattne, J, Grimshaw, S, Borek, D, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-01 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: |
|
|
2GDA
 
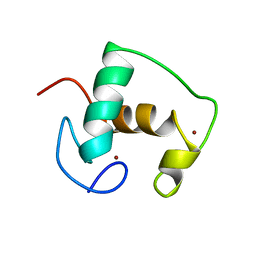 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | Deposit date: | 1994-03-15 | | Release date: | 1994-06-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
1YQC
 
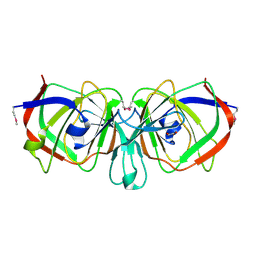 | | Crystal Structure of Ureidoglycolate Hydrolase (AllA) from Escherichia coli O157:H7 | | Descriptor: | GLYOXYLIC ACID, Ureidoglycolate hydrolase | | Authors: | Raymond, S, Tocilj, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Crystal structure of ureidoglycolate hydrolase (AllA) from Escherichia coli O157:H7
Proteins, 61, 2005
|
|
4OFD
 
 | | Crystal Structure of mouse Neph1 D1-D2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Kin of IRRE-like protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF3
 
 | | Crystal Structure of SYG-1 D1-D2, Glycosylated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFP
 
 | | Crystal Structure of SYG-2 D3-D4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-2 | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFY
 
 | | Crystal Structure of the Complex of SYG-1 D1-D2 and SYG-2 D1-D4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ETHYL MERCURY ION, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF8
 
 | | Crystal Structure of Rst D1-D2 | | Descriptor: | GLYCEROL, Irregular chiasm C-roughest protein, SODIUM ION | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF7
 
 | | Crystal Structure of SYG-1 D1, Crystal Form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFI
 
 | | Crystal Structure of Duf (Kirre) D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Kin of irre, isoform A, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF6
 
 | | Crystal Structure of SYG-1 D1, Crystal form 1 | | Descriptor: | 1,2-ETHANEDIOL, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF0
 
 | | Crystal Structure of SYG-1 D1-D2, refolded | | Descriptor: | Protein SYG-1, isoform b | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
