1VI7
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yigZ | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YIGZ, a conserved hypothetical protein from Escherichia coli k12 with a novel fold
Proteins, 55, 2004
|
|
1VIO
 
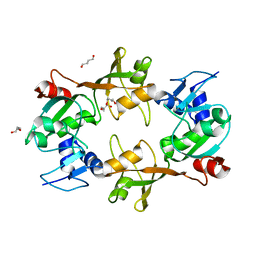 | | Crystal structure of pseudouridylate synthase | | Descriptor: | 1,4-BUTANEDIOL, Ribosomal small subunit pseudouridine synthase A | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the pseudouridine synthase RsuA from Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1VJE
 
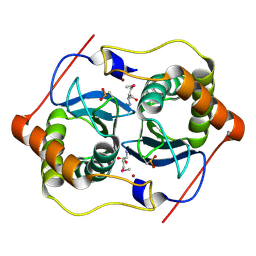 | |
3BE7
 
 | | Crystal structure of Zn-dependent arginine carboxypeptidase | | Descriptor: | ARGININE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Iizuka, M, Bain, K, Rodgers, L, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-16 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
3H14
 
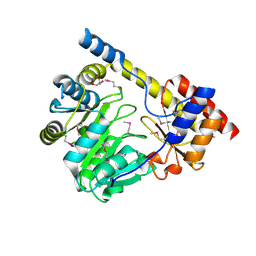 | | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, classes I and II, GLYCEROL | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Miller, S, Bain, K, Rutter, M, Tarun, G, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-05-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi
TO BE PUBLISHED
|
|
3FEQ
 
 | | Crystal structure of uncharacterized protein eah89906 | | Descriptor: | PUTATIVE AMIDOHYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Bonanno, J, Romero, R, Freeman, J, Lau, C, Smith, D, Bain, K, Wasserman, S.R, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-30 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3DUG
 
 | | Crystal structure of zn-dependent arginine carboxypeptidase complexed with zinc | | Descriptor: | ARGININE, GLYCEROL, ZINC ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Iizuka, M, Bain, K, Rodgers, L, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-17 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
