8SGT
 
 | |
8SGJ
 
 | |
8SGI
 
 | | Cryo-EM structure of human NCX1 in complex with SEA0400 | | Descriptor: | 2-{4-[(2,5-difluorophenyl)methoxy]phenoxy}-5-ethoxyaniline, CALCIUM ION, Fab heavy chain, ... | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanisms of human cardiac sodium calcium exchanger NCX1
To Be Published
|
|
3P9L
 
 | | Crystal Structure of H2-Kb in complex with the chicken ovalbumin epitope OVA | | Descriptor: | Beta-2-microglobulin, CALCIUM ION, H-2 class I histocompatibility antigen, ... | | Authors: | Wesselingh, R, Gras, S, Guillonneau, C, Turner, S.J, Rossjohn, J. | | Deposit date: | 2010-10-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses
J.Immunol., 187, 2011
|
|
6K7X
 
 | | Human MCU-EMRE complex | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhuo, W, Zhou, H, Yang, M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
6K7Y
 
 | | Intact human mitochondrial calcium uniporter complex with MICU1/MICU2 subunits | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhuo, W, Zhou, H, Yang, M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
3P9M
 
 | | Crystal Structure of H2-Kb in complex with a mutant of the chicken ovalbumin epitope OVA-G4 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Wesselingh, R, Gras, S, Guillonneau, C, Turner, S.J, Rossjohn, J. | | Deposit date: | 2010-10-18 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses
J.Immunol., 187, 2011
|
|
3PAB
 
 | | Crystal Structure of H2-Kb in complex with a mutant of the chicken ovalbumin epitope OVA-E1 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Wesselingh, R, Gras, S, Guillonneau, C, Turner, S.J, Rossjohn, J. | | Deposit date: | 2010-10-19 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses
J.Immunol., 187, 2011
|
|
4PDM
 
 | | Crystal Structure of K+ selective NaK mutant in rubidium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, RUBIDIUM ION | | Authors: | Lam, Y. | | Deposit date: | 2014-04-19 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | High Resolution Structural Views of Rubidium, Cesium and Barium Binding within a Potassium Selective Channel Filter
To Be Published
|
|
6BCJ
 
 | | cryo-EM structure of TRPM4 in apo state with short coiled coil at 3.1 angstrom resolution | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
6BCQ
 
 | | cryo-EM structure of TRPM4 in ATP bound state with long coiled coil at 3.3 angstrom resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
5YGP
 
 | | Human TNFRSF25 death domain mutant-D412E | | Descriptor: | SULFATE ION, TNFRSF25 death domain, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T.C. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
6BCO
 
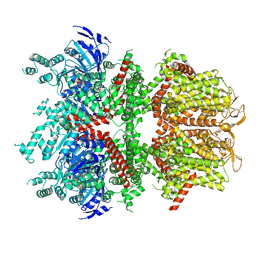 | | cryo-EM structure of TRPM4 in ATP bound state with short coiled coil at 2.9 angstrom resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
5Y2G
 
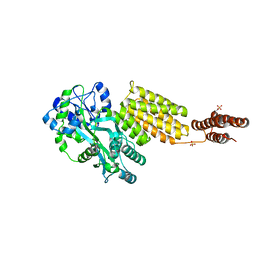 | | Structure of MBP tagged GBS CAMP | | Descriptor: | Maltose-binding periplasmic protein,Protein B, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2017-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of the CAMP factor of Streptococcus agalactiae with the aid of an MBP tag and insights into membrane-surface attachment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5YGS
 
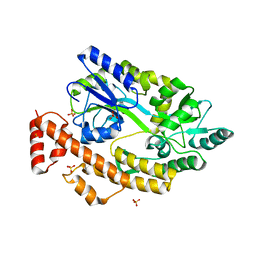 | | Human TNFRSF25 death domain | | Descriptor: | Human TNRSF25 death domain, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
6CG8
 
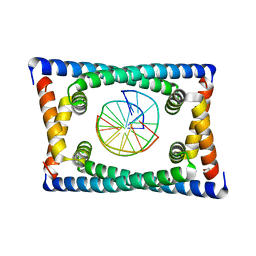 | | Structure of C. crescentus GapR-DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*AP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*TP*TP*TP*AP*A)-3'), UPF0335 protein B7Z12_12435 | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication.
Cell, 175, 2018
|
|
6CFX
 
 | | Bosea sp GapR solved in the presence of DNA | | Descriptor: | PHOSPHATE ION, UPF0335 protein ASE63_04290 | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-02-18 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication.
Cell, 175, 2018
|
|
6D80
 
 | | Cryo-EM structure of the mitochondrial calcium uniporter from N. fischeri bound to saposin | | Descriptor: | CALCIUM ION, Mitochondrial calcium uniporter, Saposin A | | Authors: | Nguyen, N.X, Armache, J.-P, Cheng, Y, Bai, X.C. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Cryo-EM structure of a fungal mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
6C9A
 
 | | Cryo-EM structure of mouse TPC1 channel in the PtdIns(3,5)P2-bound state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | She, J, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-04-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel.
Nature, 556, 2018
|
|
6D7W
 
 | | Cryo-EM structure of the mitochondrial calcium uniporter from N. fischeri at 3.8 Angstrom resolution | | Descriptor: | CALCIUM ION, Mitochondrial calcium uniporter | | Authors: | Nguyen, N.X, Armache, J.-P, Cheng, Y, Bai, X.C. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of a fungal mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
6BYJ
 
 | | Structure of human 14-3-3 gamma bound to O-GlcNAc peptide | | Descriptor: | 14-3-3 protein gamma, 2-acetamido-2-deoxy-beta-D-glucopyranose, TSTTATPPVSQASSTTTSTW O-GlcNac peptide | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
6CFY
 
 | | Bosea sp Root 381 apo GapR structure | | Descriptor: | UPF0335 protein ASE63_04290 | | Authors: | Schumacherr, M.A. | | Deposit date: | 2018-02-18 | | Release date: | 2018-09-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication.
Cell, 175, 2018
|
|
6BYL
 
 | | Structure of 14-3-3 gamma bound to O-GlcNAcylated thr peptide | | Descriptor: | 14-3-3 protein gamma, 2-acetamido-2-deoxy-beta-D-glucopyranose, TSASTTVPVTTATTTTTSTW O-GlcNac peptide | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
5ZNY
 
 | | Structure of mDR3_DD-C363G with MBP tag | | Descriptor: | Maltose-binding periplasmic protein,Tumor necrosis factor receptor superfamily, member 25, SULFATE ION | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
6BYK
 
 | | Structure of 14-3-3 beta/alpha bound to O-ClcNAc peptide | | Descriptor: | 14-3-3 protein beta/alpha, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATPPVSQASSTT O-GlcNac peptide | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
