6ZYJ
 
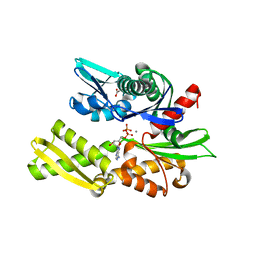 | | Crystal structure of Hsc70 ATPase domain in complex with ADP and calcium | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Yan, Y, Preissler, S, Ron, D. | | Deposit date: | 2020-08-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Calcium depletion challenges endoplasmic reticulum proteostasis by destabilising BiP-substrate complexes.
Elife, 9, 2020
|
|
6ZYH
 
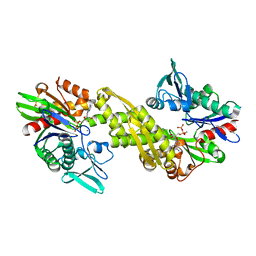 | |
3NAX
 
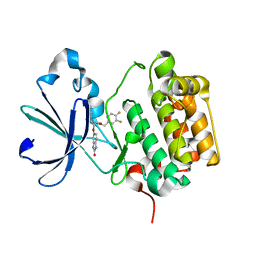 | | PDK1 in complex with inhibitor MP7 | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and pharmacological inhibition of PDK1 in cancer cells: characterization of a selective allosteric kinase inhibitor.
J.Biol.Chem., 286, 2011
|
|
3NAY
 
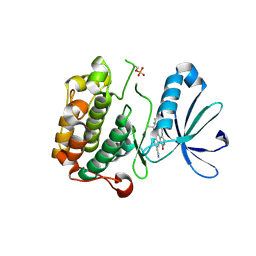 | | PDK1 in complex with inhibitor MP6 | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 4-(2-cyclopropylethylidene)-9-(1H-pyrazol-4-yl)-6-{[(1R)-1,2,2-trimethylpropyl]amino}benzo[c][1,6]naphthyridin-1(4H)-one | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective inhibition of PDK1 using an allosteric kinase inhibitor and RNAi impairs cancer cell migration and anchorage-independent growth of primary tumor lines
J.Biol.Chem., 2010
|
|
2HOG
 
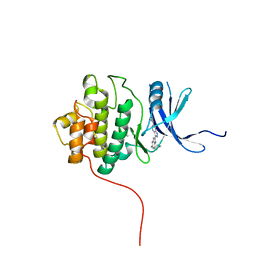 | | crystal structure of Chek1 in complex with inhibitor 20 | | Descriptor: | (5-{3-[5-(PIPERIDIN-1-YLMETHYL)-1H-INDOL-2-YL]-1H-INDAZOL-6-YL}-2H-1,2,3-TRIAZOL-4-YL)METHANOL, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Ikuta, M. | | Deposit date: | 2006-07-14 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3-(Indol-2-yl)indazoles as Chek1 kinase inhibitors: Optimization of potency and selectivity via substitution at C6.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FKY
 
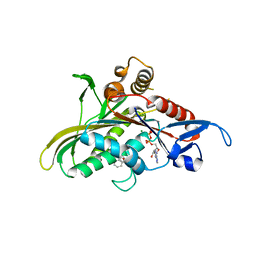 | | crystal structure of KSP in complex with inhibitor 13 | | Descriptor: | (2S)-4-(2,5-DIFLUOROPHENYL)-N-METHYL-2-PHENYL-N-PIPERIDIN-4-YL-2,5-DIHYDRO-1H-PYRROLE-1-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-01-05 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 2: the design, synthesis, and characterization of 2,4-diaryl-2,5-dihydropyrrole inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3C6T
 
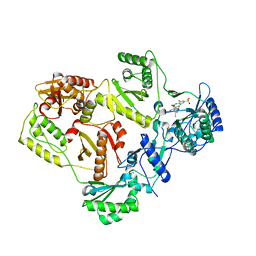 | | Crystal Structure of HIV Reverse Transcriptase in complex with inhibitor 14 | | Descriptor: | 2-[3-chloro-5-(3-chloro-5-cyanophenoxy)phenoxy]-N-(2-chloro-4-sulfamoylphenyl)acetamide, Reverse transcriptase | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The design and synthesis of diaryl ether second generation HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) with enhanced potency versus key clinical mutations.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2FL2
 
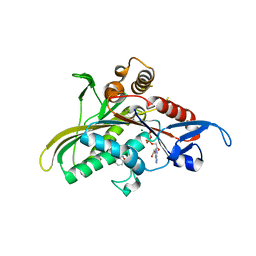 | | crystal structure of KSP in complex with inhibitor 19 | | Descriptor: | (1S)-1-CYCLOPROPYL-2-[(2S)-4-(2,5-DIFLUOROPHENYL)-2-PHENYL-2,5-DIHYDRO-1H-PYRROL-1-YL]-2-OXOETHANAMINE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-01-05 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 2: the design, synthesis, and characterization of 2,4-diaryl-2,5-dihydropyrrole inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FL6
 
 | | crystal structure of KSP in complex with inhibitor 6 | | Descriptor: | (2S)-4-(2,5-DIFLUOROPHENYL)-N,N-DIMETHYL-2-PHENYL-2,5-DIHYDRO-1H-PYRROLE-1-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-01-05 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 2: the design, synthesis, and characterization of 2,4-diaryl-2,5-dihydropyrrole inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3I0R
 
 | | crystal structure of HIV reverse transcriptase in complex with inhibitor 3 | | Descriptor: | Reverse transcriptase/ribonuclease H, S-{2-[(2-chloro-4-sulfamoylphenyl)amino]-2-oxoethyl} 6-methyl-3,4-dihydroquinoline-1(2H)-carbothioate, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Substituted tetrahydroquinolines as potent allosteric inhibitors of reverse transcriptase and its key mutants.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3I0S
 
 | | crystal structure of HIV reverse transcriptase in complex with inhibitor 7 | | Descriptor: | Reverse transcriptase/ribonuclease H, S-{2-[(2-chloro-4-sulfamoylphenyl)amino]-2-oxoethyl} 6,8-dichloro-3,4-dihydroquinoline-1(2H)-carbothioate, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substituted tetrahydroquinolines as potent allosteric inhibitors of reverse transcriptase and its key mutants.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4AJT
 
 | |
3F9N
 
 | | Crystal structure of chk1 kinase in complex with inhibitor 38 | | Descriptor: | 3-(3-chlorophenyl)-2-({(1S)-1-[(6S)-2,8-diazaspiro[5.5]undec-2-ylcarbonyl]pentyl}sulfanyl)quinazolin-4(3H)-one, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S, Ikuta, M. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of thioquinazolinones, allosteric Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3CJO
 
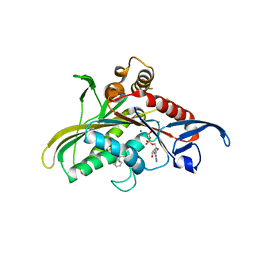 | | Crystal structure of KSP in complex with inhibitor 30 | | Descriptor: | (2S)-4-(2,5-difluorophenyl)-N-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2008-03-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. 9. Discovery of (2S)-4-(2,5-difluorophenyl)-n-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide (MK-0731) for the treatment of taxane-refractory cancer.
J.Med.Chem., 51, 2008
|
|
2RF2
 
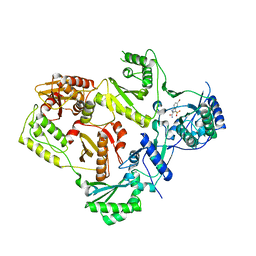 | | HIV reverse transcriptase in complex with inhibitor 7e (NNRTI) | | Descriptor: | 5-bromo-3-(pyrrolidin-1-ylsulfonyl)-1H-indole-2-carboxamide, Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC 2.7.7.7) (EC 3.1.26.4) (p66 RT) | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2007-09-27 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel indole-3-sulfonamides as potent HIV non-nucleoside reverse transcriptase inhibitors (NNRTIs).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3T1A
 
 | |
3T19
 
 | |
2Q2Z
 
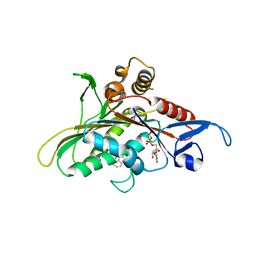 | | Crystal Structure of KSP in Complex with Inhibitor 22 | | Descriptor: | 1-[(4R)-4-[3-(4-ACETYLPIPERAZIN-1-YL)PROPYL]-1-(2-FLUORO-5-METHYLPHENYL)-4-PHENYL-4,5-DIHYDRO-1H-PYRAZOL-3-YL]ETHANONE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2007-05-29 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 8: Design and synthesis of 1,4-diaryl-4,5-dihydropyrazoles as potent inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
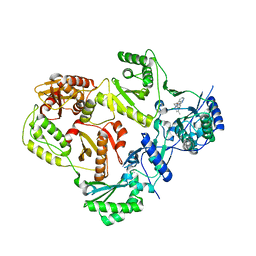
3TAM
 
 | |
2Q2Y
 
 | | Crystal Structure of KSP in complex with Inhibitor 1 | | Descriptor: | 1-{(3R,3AR)-3-[3-(4-ACETYLPIPERAZIN-1-YL)PROPYL]-8-FLUORO-3-PHENYL-3A,4-DIHYDRO-3H-PYRAZOLO[5,1-C][1,4]BENZOXAZIN-2-YL}ETHANONE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2007-05-29 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 8: Design and synthesis of 1,4-diaryl-4,5-dihydropyrazoles as potent inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2QHM
 
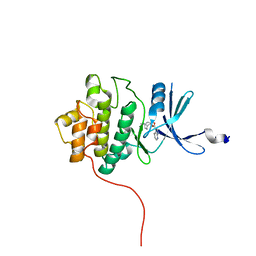 | | crystal structure of Chek1 in complex with inhibitor 2a | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLATE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S. | | Deposit date: | 2007-07-02 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of a pyrazoloquinolinone class of Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3LP3
 
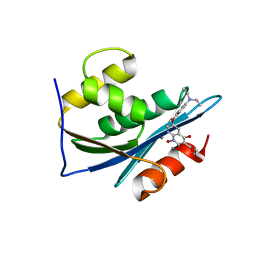 | | p15 HIV RNaseH domain with inhibitor MK3 | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, p15 | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3LP2
 
 | | HIV-1 reverse transcriptase with inhibitor | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3LP1
 
 | | HIV-1 reverse transcriptase with inhibitor | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 3-cyclopentyl-1,4-dihydroxy-1,8-naphthyridin-2(1H)-one, MANGANESE (II) ION, ... | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3LP0
 
 | | HIV-1 reverse transcriptase with inhibitor | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
