1J8M
 
 | | Signal Recognition Particle conserved GTPase domain from A. ambivalens | | Descriptor: | SIGNAL RECOGNITION 54 KDA PROTEIN | | Authors: | Montoya, G, te Kaat, K, Moll, R, Schafer, G, Sinning, I. | | Deposit date: | 2001-05-22 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the conserved GTPase of SRP54 from the archaeon Acidianus ambivalens and its comparison with related structures suggests a model for the SRP-SRP receptor complex.
Structure Fold.Des., 8, 2000
|
|
1MHN
 
 | |
3MC9
 
 | |
3MC8
 
 | |
3UI2
 
 | |
4ADU
 
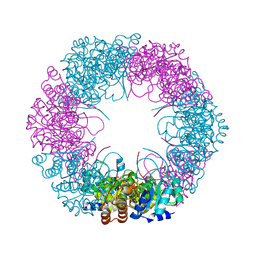 | | Crystal structure of plasmodial PLP synthase with bound R5P intermediate | | Descriptor: | 1,2-ETHANEDIOL, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, PUTATIVE, ... | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
4ADS
 
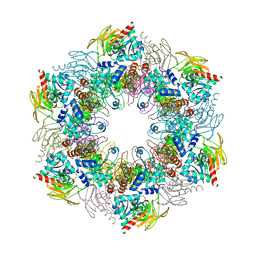 | | Crystal structure of plasmodial PLP synthase complex | | Descriptor: | PDX2 PROTEIN, PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, ... | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
4ADT
 
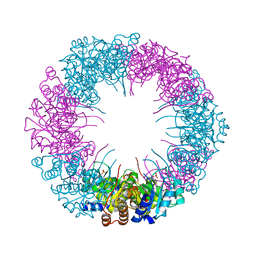 | | Crystal structure of plasmodial PLP synthase | | Descriptor: | PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, PUTATIVE | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
3BB3
 
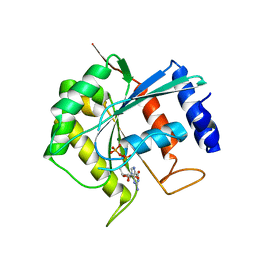 | | Crystal structure of Toc33 from Arabidopsis thaliana in complex with GDP and Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 protein | | Authors: | Koenig, P, Sinning, I, Schleiff, E, Tews, I. | | Deposit date: | 2007-11-09 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The GTPase cycle of the chloroplast import receptors Toc33/Toc34: implications from monomeric and dimeric structures.
Structure, 16, 2008
|
|
3BB4
 
 | | Crystal structure of Toc33 from Arabidopsis thaliana in complex with Mg2+ and GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, T7I23.11 protein | | Authors: | Koenig, P, Sinning, I, Schleiff, E, Tews, I. | | Deposit date: | 2007-11-09 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The GTPase cycle of the chloroplast import receptors Toc33/Toc34: implications from monomeric and dimeric structures.
Structure, 16, 2008
|
|
3DEF
 
 | | Crystal structure of Toc33 from Arabidopsis thaliana, dimerization deficient mutant R130A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 protein | | Authors: | Koenig, P, Schleiff, E, Sinning, I, Tews, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | On the significance of Toc-GTPase homodimers
J.Biol.Chem., 283, 2008
|
|
3DEP
 
 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
3DWV
 
 | |
3FEM
 
 | |
2ABW
 
 | | Glutaminase subunit of the plasmodial PLP synthase (Vitamin B6 biosynthesis) | | Descriptor: | Pdx2 protein, TETRAETHYLENE GLYCOL | | Authors: | Gengenbacher, M, Fitzpatrick, T.B, Raschle, T, Flicker, K, Sinning, I, Mueller, S, Macheroux, P, Tews, I, Kappes, B. | | Deposit date: | 2005-07-17 | | Release date: | 2006-01-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Vitamin B6 Biosynthesis by the Malaria Parasite Plasmodium falciparum: Biochemical and structural insights
J.Biol.Chem., 281, 2006
|
|
6Y31
 
 | | NG domain of human SRP54 T117 deletion mutant | | Descriptor: | Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Lapouge, K, Becker, M.M.M, Kotova, I, Haas, M, Carapito, R, Wild, K, Bahram, S, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
3D8E
 
 | |
3D8F
 
 | | Crystal structure of the human Fe65-PTB1 domain with bound phosphate (trigonal crystal form) | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1, PHOSPHATE ION | | Authors: | Radzimanowski, J, Ravaud, S, Sinning, I, Wild, K. | | Deposit date: | 2008-05-23 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the human Fe65-PTB1 domain.
J.Biol.Chem., 283, 2008
|
|
3D8D
 
 | | Crystal structure of the human Fe65-PTB1 domain | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 precursor protein-binding family B member 1, MERCURY (II) ION | | Authors: | Radzimanowski, J, Ravaud, S, Sinning, I, Wild, K. | | Deposit date: | 2008-05-23 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human Fe65-PTB1 domain.
J.Biol.Chem., 283, 2008
|
|
3DXE
 
 | |
3DXC
 
 | |
3DXD
 
 | |
5TKY
 
 | | Crystal structure of the co-translational Hsp70 chaperone Ssb in the ATP-bound, open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Gumiero, A, Gese, G.V, Weyer, F.A, Lapouge, K, Sinning, I. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interaction of the cotranslational Hsp70 Ssb with ribosomal proteins and rRNA depends on its lid domain.
Nat Commun, 7, 2016
|
|
2EV4
 
 | | Structure of Rv1264N, the regulatory domain of the mycobacterial adenylyl cylcase Rv1264, with a salt precipitant | | Descriptor: | CHLORIDE ION, Hypothetical protein Rv1264/MT1302, OLEIC ACID | | Authors: | Findeisen, F, Tews, I, Sinning, I. | | Deposit date: | 2005-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid
J.Mol.Biol., 369, 2007
|
|
2EV2
 
 | | Structure of Rv1264N, the regulatory domain of the mycobacterial adenylyl cylcase Rv1264, at pH 8.5 | | Descriptor: | Hypothetical protein Rv1264/MT1302, OLEIC ACID | | Authors: | Findeisen, F, Tews, I, Sinning, I. | | Deposit date: | 2005-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid
J.Mol.Biol., 369, 2007
|
|
