1LY4
 
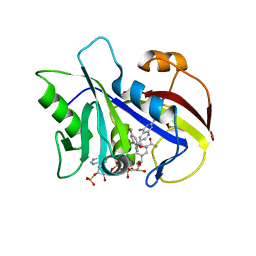 | | Analysis of quinazoline and PYRIDO[2,3D]PYRIMIDINE N9-C10 reversed bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LY3
 
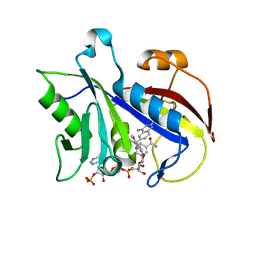 | | ANALYSIS OF QUINAZOLINE AND PYRIDOPYRIMIDINE N9-C10 REVERSED BRIDGE ANTIFOLATES IN COMPLEX WITH NADP+ AND PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE | | Descriptor: | 2,4-DIAMINO-6-[N-(2',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]QUINAZOLINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1P49
 
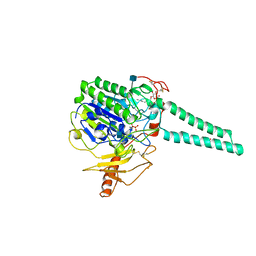 | | Structure of Human Placental Estrone/DHEA Sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Hernandez-Guzman, F.G, Higashiyama, T, Pangborn, W, Osawa, Y, Ghosh, D. | | Deposit date: | 2003-04-21 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Human Estrone Sulfatase Suggests Functional Roles of Membrane Association
J.Biol.Chem., 278, 2003
|
|
1PD8
 
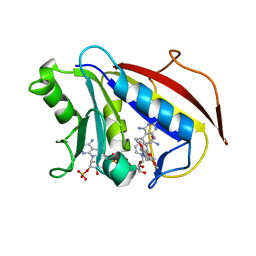 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine Antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-5-METHYL-6-[(3,4,5-TRIMETHOXY-N-METHYLANILINO)METHYL]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PDB
 
 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine Antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | Dihydrofolate reductase | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PD9
 
 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-5-METHYL-6-[(3,4,5-TRIMETHOXY-N-METHYLANILINO)METHYL]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4CD2
 
 | | LIGAND INDUCED CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURES OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COMPLEXES WITH FOLATE AND NADP+ | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID | | Authors: | Cody, V, Galitsky, N, Rak, D, Luft, J.R, Pangborn, W, Queener, S.F. | | Deposit date: | 1999-03-18 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and NADP+.
Biochemistry, 38, 1999
|
|
7CLI
 
 | | Structure of NF-kB p52 homodimer bound to P-Selectin kB DNA fragment | | Descriptor: | DNA (5'-D(*CP*AP*AP*GP*GP*GP*GP*TP*CP*AP*CP*CP*CP*CP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*GP*GP*GP*GP*GP*TP*GP*AP*CP*CP*CP*CP*TP*TP*G)-3'), Nuclear factor NF-kappa-B p52 subunit | | Authors: | Meshcheryakov, V.A, Wang, V.Y.-F. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of NF-kappa B p52 homodimer-DNA complexes rationalize binding mechanisms and transcription activation.
Elife, 12, 2023
|
|
7VUP
 
 | |
7VUQ
 
 | |
7W7L
 
 | |
7RML
 
 | | Neisseria meningitidis Methylenetetrahydrofolate reductase in complex with FAD | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pederick, J.L, Wegener, K.L, Salaemae, W, Bruning, J.B. | | Deposit date: | 2021-07-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural characterization of meningococcal methylenetetrahydrofolate reductase.
Protein Sci., 32, 2023
|
|
3LON
 
 | | HCV NS3-4a protease domain with ketoamide inhibitor narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, BETA-MERCAPTOETHANOL, Genome polyprotein, ... | | Authors: | Prongay, A.J. | | Deposit date: | 2010-02-04 | | Release date: | 2011-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Candidate selection and preclinical evaluation culminating in the discovery of Narlaprevir (SCH 900518): A potent, selective and orally efficacious second generation HCV NS3 serine protease inhibitor
To be Published
|
|
7UP2
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP1
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP3
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (3P)-4-[4-(hydroxymethyl)phenyl]-3-(2H-tetrazol-5-yl)pyridine-2-sulfonamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOX
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-(hydroxymethyl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-ol, ACETATE ION, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOY
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (6P)-4-amino-6-(2H-tetrazol-5-yl)benzene-1,3-disulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
3LOX
 
 | | HCV NS3-4a protease domain with a ketoamide inhibitor derivative of Boceprevir bound | | Descriptor: | (1R,2S,5S)-N-[(2S,3R)-4-amino-1-cyclobutyl-3-hydroxy-4-oxobutan-2-yl]-6,6-dimethyl-3-{3-methyl-N-[(1-methylcyclohexyl)c arbamoyl]-L-valyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, BETA-MERCAPTOETHANOL, HCV NS3 Protease, ... | | Authors: | Prongay, A.J. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The introduction of P4 substituted 1-methylcyclohexyl groups into Boceprevir: a change in direction in the search for a second generation HCV NS3 protease inhibitor.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5HMK
 
 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {4-[2-(2-hydroxyethoxy)phenyl]piperazin-1-yl}[(2R,3S)-2-propyl-3-[4-(trifluoromethyl)phenoxy]-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}piperidin-3-yl]methanone | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HMI
 
 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, {4-[2-(2-hydroxyethoxy)phenyl]piperazin-1-yl}[(2R,3S)-2-propyl-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}-3-{[5-(trifluoromethyl)thiophen-3-yl]oxy}piperidin-3-yl]methanone | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HMH
 
 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | 4-[2-(4-{[(2R,3S)-2-propyl-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}-3-{[5-(trifluoromethyl)thiophen-3-yl]oxy}piperidin-3-yl]carbonyl}piperazin-1-yl)phenoxy]butanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
1HDC
 
 | |
6LYW
 
 | | Structural insight into the biological functions of Arabidopsis thaliana ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
6LYX
 
 | | Crystal structure of oxidized ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Cai, W.G, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
