3LEA
 
 | |
3LGP
 
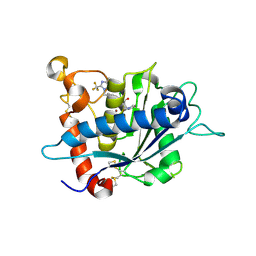 | | Crystal structure of catalytic domain of tace with benzimidazolyl-thienyl-tartrate based inhibitor | | Descriptor: | (2R,3R)-4-[(2R)-2-(3-chlorophenyl)pyrrolidin-1-yl]-2,3-dihydroxy-4-oxo-N-[(5-{[2-(trifluoromethyl)-1H-benzimidazol-1-yl]methyl}thiophen-2-yl)methyl]butanamide, Disintegrin and metalloproteinase domain-containing protein 17, ZINC ION | | Authors: | Orth, P. | | Deposit date: | 2010-01-21 | | Release date: | 2010-07-28 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and activity relationships of tartrate-based TACE inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LNK
 
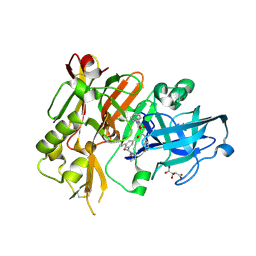 | | Structure of BACE bound to SCH743813 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylcarbonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Orth, P, Cumming, J. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LE9
 
 | |
3E8R
 
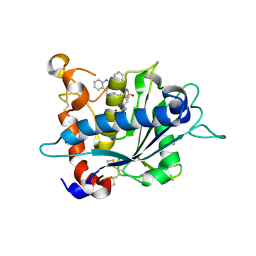 | | Crystal structure of catalytic domain of TACE with hydroxamate inhibitor | | Descriptor: | (1R,2S)-N~2~-hydroxy-1-{4-[(2-phenylquinolin-4-yl)methoxy]benzyl}cyclopropane-1,2-dicarboxamide, ADAM 17, CITRIC ACID, ... | | Authors: | Orth, P. | | Deposit date: | 2008-08-20 | | Release date: | 2008-10-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel hydroxamates as highly potent tumor necrosis factor-alpha converting enzyme inhibitors. Part II: optimization of the S3' pocket.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3EWJ
 
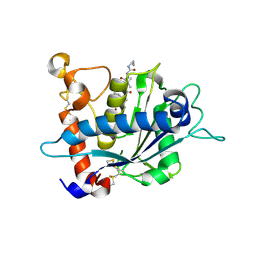 | | Crystal structure of catalytic domain of TACE with carboxylate inhibitor | | Descriptor: | (1S,3R,6S)-4-oxo-6-{4-[(2-phenylquinolin-4-yl)methoxy]phenyl}-5-azaspiro[2.4]heptane-1-carboxylic acid, ADAM 17, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Orth, P. | | Deposit date: | 2008-10-15 | | Release date: | 2008-11-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of novel spirocyclopropyl hydroxamate and carboxylate compounds as TACE inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2FJ1
 
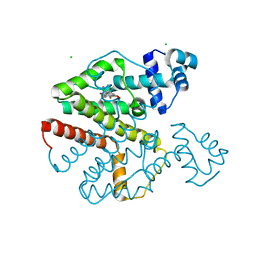 | | Crystal Structure Analysis of Tet Repressor (class D) in Complex with 7-Chlortetracycline-Nickel(II) | | Descriptor: | 7-CHLOROTETRACYCLINE, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Orth, P, Saenger, W, Palm, G.J, Hinrichs, W. | | Deposit date: | 2005-12-30 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Specific binding of divalent metal ions to tetracycline and to the Tet repressor/tetracycline complex.
J.Biol.Inorg.Chem., 13, 2008
|
|
2FV5
 
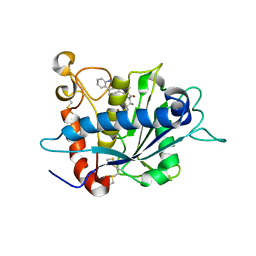 | | Crystal structure of TACE in complex with IK682 | | Descriptor: | (2R)-N-HYDROXY-2-[(3S)-3-METHYL-3-{4-[(2-METHYLQUINOLIN-4-YL)METHOXY]PHENYL}-2-OXOPYRROLIDIN-1-YL]PROPANAMIDE, ADAM 17, ZINC ION | | Authors: | Orth, P, Niu, X. | | Deposit date: | 2006-01-30 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IK682, a tight binding inhibitor of TACE.
Arch.Biochem.Biophys., 451, 2006
|
|
2FV9
 
 | | Crystal structure of TACE in complex with JMV 390-1 | | Descriptor: | ADAM 17, N-[(2R)-2-BENZYL-4-(HYDROXYAMINO)-4-OXOBUTANOYL]-L-ISOLEUCYL-L-LEUCINE, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Orth, P. | | Deposit date: | 2006-01-30 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Stabilization of the autoproteolysis of TNF-alpha converting enzyme (TACE) results in a novel crystal form suitable for structure-based drug design studies.
Protein Eng.Des.Sel., 19, 2006
|
|
4MC2
 
 | | HIV protease in complex with SA525P | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4R)-4-tert-butyl-7-fluoro-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
4MC6
 
 | | HIV protease in complex with SA499 | | Descriptor: | 1,2-ETHANEDIOL, 1-tert-butyl-3-{(2S,3R)-4-[(4R)-7-fluoro-1,1-dioxido-4-(propan-2-yl)-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}urea, CHLORIDE ION, ... | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
1ILX
 
 | | Excited State Dynamics in Photosystem II Revised. New Insights from the X-ray Structure. | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, CADMIUM ION, CHLOROPHYLL A, ... | | Authors: | Vasilev, S, Orth, P, Zouni, A, Owens, T.G, Bruce, D. | | Deposit date: | 2001-05-09 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Excited-state dynamics in photosystem II: insights from the x-ray crystal structure.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IRQ
 
 | | Crystal structure of omega transcriptional repressor at 1.5A resolution | | Descriptor: | omega transcriptional repressor | | Authors: | Murayama, K, Orth, P, De La Hoz, A.B, Alonso, J.C, Saenger, W. | | Deposit date: | 2001-10-11 | | Release date: | 2001-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of omega transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 A resolution.
J.Mol.Biol., 314, 2001
|
|
4MC1
 
 | | HIV protease in complex with SA526P | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4S)-4-tert-butyl-7-fluoro-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
4MC9
 
 | | HIV protease in complex with AA74 | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4R)-7-fluoro-1,1-dioxido-4-(propan-2-yl)-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
8TX6
 
 | |
8TX5
 
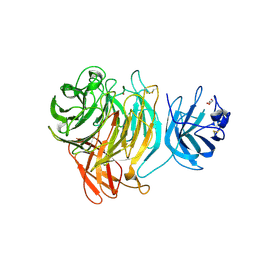 | |
3JZI
 
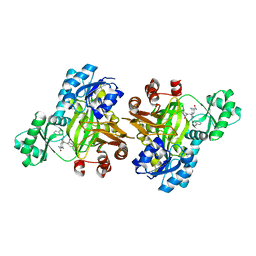 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EDZ
 
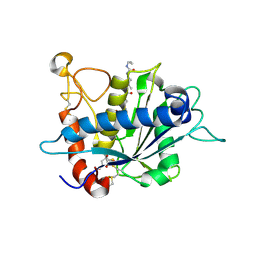 | | Crystal structure of catalytic domain of TACE with hydroxamate inhibitor | | Descriptor: | ADAM 17, CITRIC ACID, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Mazzola, R.D, Zhu, Z, Sinning, L, McKittrick, B, Lavey, B, Spitler, J, Kozlowski, J, Neng-Yang, S, Zhou, G, Guo, Z, Orth, P, Madison, V, Sun, J, Lundell, D, Niu, X. | | Deposit date: | 2008-09-03 | | Release date: | 2008-09-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel hydroxamates as highly potent tumor necrosis factor-alpha converting enzyme inhibitors. Part II: optimization of the S3' pocket.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1FE1
 
 | | CRYSTAL STRUCTURE PHOTOSYSTEM II | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, CADMIUM ION, CHLOROPHYLL A, ... | | Authors: | Zouni, A, Witt, H.-T, Kern, J, Fromme, P, Krauss, N, Saenger, W, Orth, P. | | Deposit date: | 2000-07-20 | | Release date: | 2001-02-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution.
Nature, 409, 2001
|
|
1A9E
 
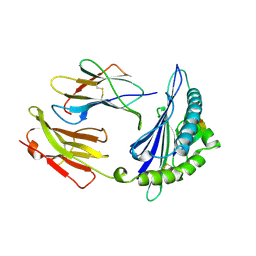 | | DECAMER-LIKE CONFORMATION OF A NANO-PEPTIDE BOUND TO HLA-B3501 DUE TO NONSTANDARD POSITIONING OF THE C-TERMINUS | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-35 B*3501 (ALPHA CHAIN), ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1998-04-05 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus.
J.Mol.Biol., 285, 1999
|
|
1A9B
 
 | | DECAMER-LIKE CONFORMATION OF A NANO-PEPTIDE BOUND TO HLA-B3501 DUE TO NONSTANDARD POSITIONING OF THE C-TERMINUS | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-35 B*3501 (ALPHA CHAIN), ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1998-04-03 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus.
J.Mol.Biol., 285, 1999
|
|
1CH0
 
 | | RNASE T1 VARIANT WITH ALTERED GUANINE BINDING SEGMENT | | Descriptor: | CALCIUM ION, CHLORIDE ION, GUANOSINE-2'-MONOPHOSPHATE, ... | | Authors: | Hoeschler, K, Hoier, H, Orth, P, Hubner, B, Saenger, W, Hahn, U. | | Deposit date: | 1999-03-30 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of an RNase T1 variant with an altered guanine binding segment.
J.Mol.Biol., 294, 1999
|
|
1CG9
 
 | | COMPLEX RECOGNITION OF THE SUPERTYPIC BW6-DETERMINANT ON HLA-B AND-C MOLECULES BY THE MONOCLONAL ANTIBODY SFR8-B6 | | Descriptor: | PROTEIN (BETA-2-MICROGLOBULIN), PROTEIN (EBNA-6 NUCLEAR PROTEIN (EBNA-3C) (EBNA-4B)), PROTEIN (HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1999-03-25 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Recognition of the Supertypic Bw6-Determinant on HLA-B and-C Molecules
by the Monoclonal Antibody SFR8-B6
To be Published
|
|
2XSE
 
 | | The structural basis for recognition of J-base containing DNA by a novel DNA-binding domain in JBP1 | | Descriptor: | GLYCEROL, NITRATE ION, THYMINE DIOXYGENASE JBP1 | | Authors: | Heidebrecht, T, Christodoulou, E, Chalmers, M.J, Jan, S, ter Riete, B, Grover, R.K, Joosten, R.P, Littler, D, vanLuenen, H, Griffin, P.R, Wentworth, P, Borst, P, Perrakis, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-03-30 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Recognition of Base J Containing DNA by a Novel DNA Binding Domain in Jbp1.
Nucleic Acids Res., 39, 2011
|
|
