7V93
 
 | | Cryo-EM structure of the Cas12c2-sgRNA binary complex | | Descriptor: | cas12c2, sgRNA | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
7V94
 
 | | Cryo-EM structure of the Cas12c2-sgRNA-target DNA ternary complex | | Descriptor: | Cas12c2, sgRNA, target DNA (non target strand), ... | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
7ETP
 
 | | Crystal structure of Pro-Phe-Leu-Phe | | Descriptor: | Pro-Phe-Leu-Phe | | Authors: | Kurumida, Y, Ikeda, K, Nakamichi, Y, Hirano, A, Kobayashi, K, Saito, Y, Kameda, T. | | Deposit date: | 2021-05-13 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.09488 Å) | | Cite: | Crystal structure of Pro-Phe-Leu-Phe
To Be Published
|
|
7ETQ
 
 | | Crystal structure of Pro-Met-Leu-Leu | | Descriptor: | Pro-Met-Leu-Leu | | Authors: | Kurumida, Y, Ikeda, K, Nakamichi, Y, Hirano, A, Kobayashi, K, Saito, Y, Kameda, T. | | Deposit date: | 2021-05-13 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structure of Pro-Met-Leu-Leu
To Be Published
|
|
7ETN
 
 | | Crystal structure of Pro-Phe-Leu-Ile | | Descriptor: | PRO-PHE-LEU-ILE | | Authors: | Kurumida, Y, Ikeda, K, Nakamichi, Y, Hirano, A, Kobayashi, K, Saito, Y, Kameda, T. | | Deposit date: | 2021-05-13 | | Release date: | 2022-05-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (0.82 Å) | | Cite: | Crystal structure of Pro-Phe-Leu-Ile
To Be Published
|
|
3VGD
 
 | | Ctystal structure of glycosyltrehalose trehalohydrolase (D252E) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGF
 
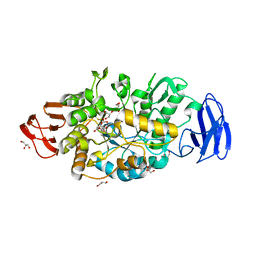 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) complexed with maltotriosyltrehalose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGH
 
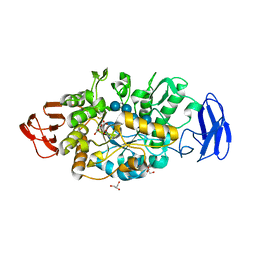 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltotriosyltrehalose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGG
 
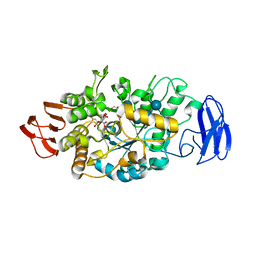 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltoheptaose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGB
 
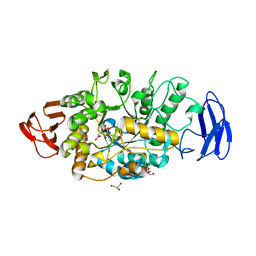 | | Crystal structure of glycosyltrehalose trehalohydrolase (GTHase) from Sulfolobus solfataricus KM1 | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGE
 
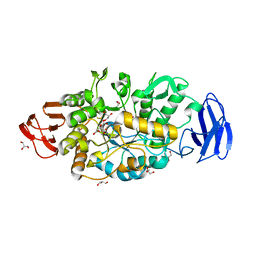 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
7D3S
 
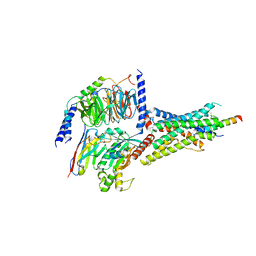 | | Human SECR in complex with an engineered Gs heterotrimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Fukuhara, S, Kobayashi, K, Kusakizako, T, Shihoya, W, Nureki, O. | | Deposit date: | 2020-09-20 | | Release date: | 2020-11-04 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the human secretin receptor coupled to an engineered heterotrimeric G protein.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
3WUH
 
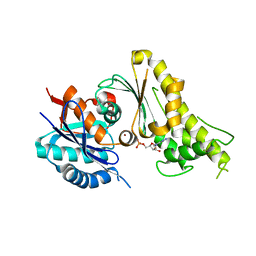 | | Qri7 and AMP complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ZINC ION, tRNA N6-adenosine threonylcarbamoyltransferase, ... | | Authors: | Tominaga, T, Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-24 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.937 Å) | | Cite: | Structure of Saccharomyces cerevisiae mitochondrial Qri7 in complex with AMP
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
1V4R
 
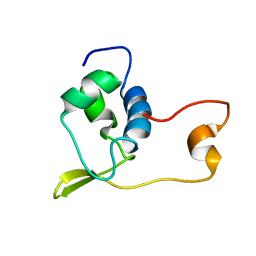 | | Solution structure of Streptmycal repressor TraR | | Descriptor: | Transcriptional Repressor | | Authors: | Tanaka, T, Komatsu, C, Kobayashi, K, Sugai, M, Kataoka, M, Kohno, T. | | Deposit date: | 2003-11-17 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Streptmycal repressor TraR
TO BE PUBLISHED
|
|
2ZHG
 
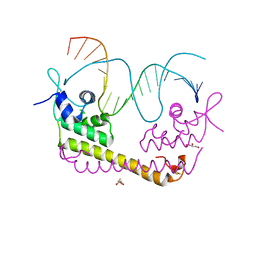 | | Crystal structure of SoxR in complex with DNA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA (5'-D(*DGP*DCP*DCP*DTP*DCP*DAP*DAP*DGP*DTP*DTP*DAP*DAP*DCP*DTP*DTP*DGP*DAP*DGP*DGP*DC)-3'), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZHH
 
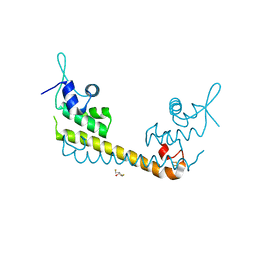 | | Crystal structure of SoxR | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FE2/S2 (INORGANIC) CLUSTER, Redox-sensitive transcriptional activator soxR | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3AQ8
 
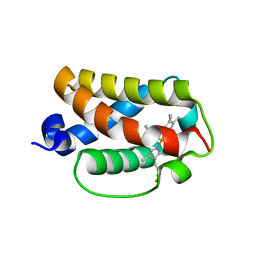 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Q46E mutant, Fe(III) form | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
3AQ9
 
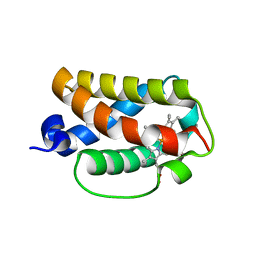 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Q50E mutant, Fe(III) form | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
3AQ6
 
 | |
3AQ7
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Y25F mutant, Fe(III) form | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
3AQ5
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Fe(II)-O2 form | | Descriptor: | Group 1 truncated hemoglobin, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
4WY3
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3R,4aS,8aR)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-11-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4TWW
 
 | | Structure of SARS-3CL protease complex with a Bromobenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(4-bromobenzoyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TWY
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
