3IZ3
 
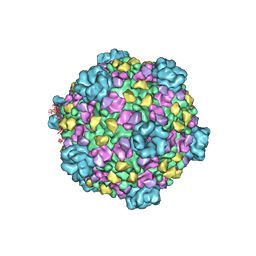 | | CryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Structural protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Cheng, L, Sun, J, Zhang, K, Mou, Z, Huang, X, Ji, G, Sun, F, Zhang, J, Zhu, P. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4I6P
 
 | | Crystal structure of Par3-NTD domain | | Descriptor: | Partitioning defective 3 homolog | | Authors: | Wang, W, Gao, F, Gong, W, Sun, F, Feng, W. | | Deposit date: | 2012-11-29 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the intrinsic self-assembly of par-3 N-terminal domain.
Structure, 21, 2013
|
|
7VOP
 
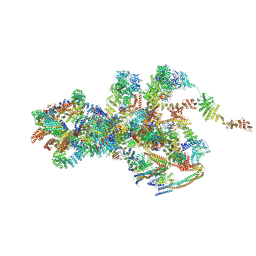 | | Cryo-EM structure of Xenopus laevis nuclear pore complex cytoplasmic ring subunit | | Descriptor: | GATOR complex protein SEC13, IL4I1 protein, MGC154553 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7VCI
 
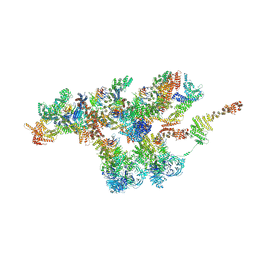 | | Structure of Xenopus laevis NPC nuclear ring asymmetric unit | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
3J1P
 
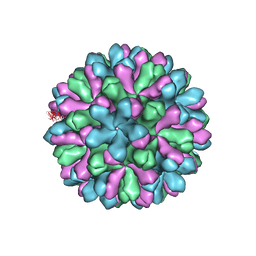 | | Atomic model of rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Wang, X, Liu, Y, Sun, F. | | Deposit date: | 2012-04-09 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
4EGT
 
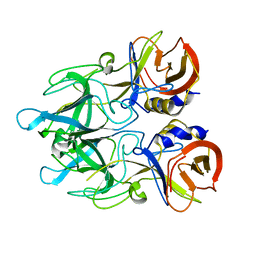 | | Crystal structure of major capsid protein P domain from rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Wang, X, Xu, F, Zhang, K, Zhai, Y, Sun, F. | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
5H3D
 
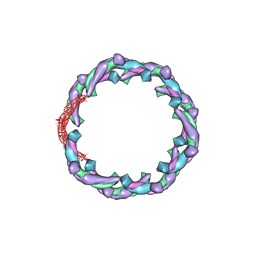 | | Helical structure of membrane tubules decorated by ACAP1 (BARPH doamin) protein by cryo-electron microscopy and MD simulation | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | Authors: | Chan, C, Pang, X.Y, Zhang, Y, Sun, F, Fan, J. | | Deposit date: | 2016-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | ACAP1 assembles into an unusual protein lattice for membrane deformation through multiple stages.
Plos Comput.Biol., 15, 2019
|
|
5J8V
 
 | |
4EJR
 
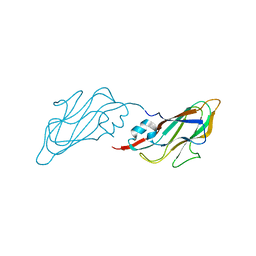 | | Crystal structure of major capsid protein S domain from rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Xu, F, Ma, J, Zhang, K, Wang, X, Sun, F. | | Deposit date: | 2012-04-07 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
7VDE
 
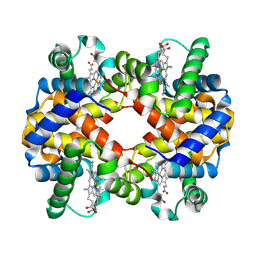 | | 3.6 A structure of the human hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, H.C, Zhang, Y, Sun, F. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI.
Nat Commun, 12, 2021
|
|
7VDA
 
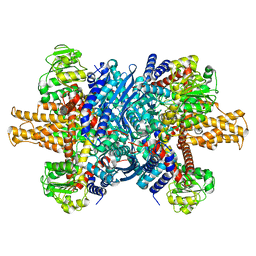 | |
7VDC
 
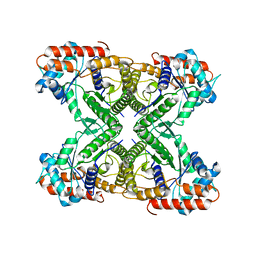 | |
7VD8
 
 | |
7VDF
 
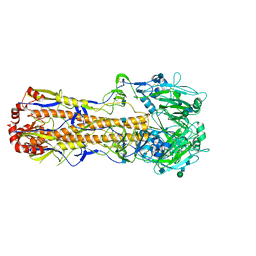 | |
7VD9
 
 | | 2.29 A structure of the human catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, H.C, Zhang, Y, Sun, F. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI.
Nat Commun, 12, 2021
|
|
2R2J
 
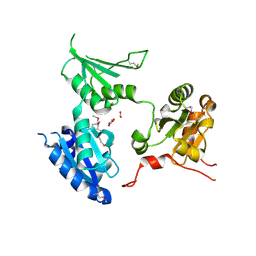 | | crystal structure of human ERp44 | | Descriptor: | FORMIC ACID, SUCCINIC ACID, Thioredoxin domain-containing protein 4 | | Authors: | Wang, L.K, Li, S.J, Sun, F, Wang, C.C. | | Deposit date: | 2007-08-25 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail.
Embo Rep., 2008
|
|
4HNE
 
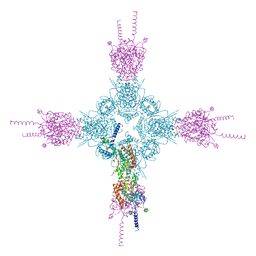 | | Crystal structure of the catalytic domain of human type II alpha Phosphatidylinositol 4-kinase (PI4KIIalpha) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
4J0E
 
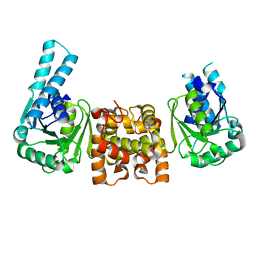 | |
4J0F
 
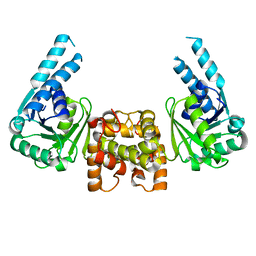 | |
3KH8
 
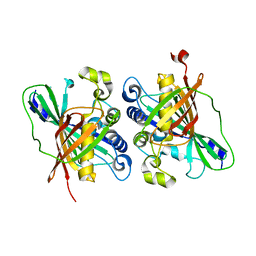 | | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici | | Descriptor: | MaoC-like dehydratase | | Authors: | Wang, H, Zhang, K, Guo, J, Zhou, Q, Zheng, X, Sun, F, Pang, H, Zhang, X. | | Deposit date: | 2009-10-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici
To be Published
|
|
7C53
 
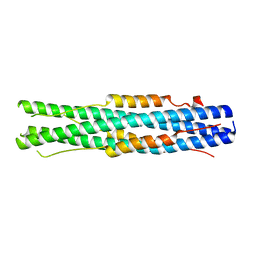 | |
3J1E
 
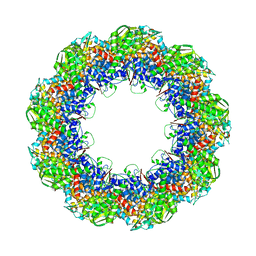 | | Cryo-EM structure of 9-fold symmetric rATcpn-beta in apo state | | Descriptor: | Chaperonin beta subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3J1B
 
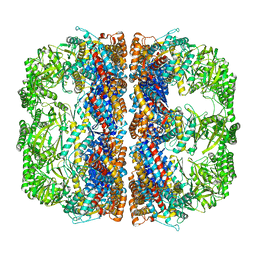 | | Cryo-EM structure of 8-fold symmetric rATcpn-alpha in apo state | | Descriptor: | Chaperonin alpha subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3J1C
 
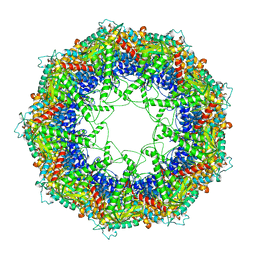 | | Cryo-EM structure of 9-fold symmetric rATcpn-alpha in apo state | | Descriptor: | Chaperonin alpha subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3K6J
 
 | | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans | | Descriptor: | PHOSPHATE ION, Protein F01G10.3, confirmed by transcript evidence, ... | | Authors: | Ouyang, Z, Zhang, K, Zhai, Y, Lu, J, Sun, F. | | Deposit date: | 2009-10-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans
To be Published
|
|
