3V0D
 
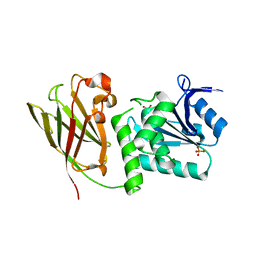 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S) | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V0G
 
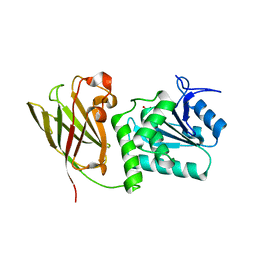 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), form III | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V0E
 
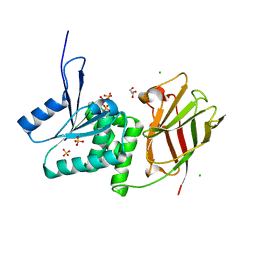 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 256-576(C363S) | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V0H
 
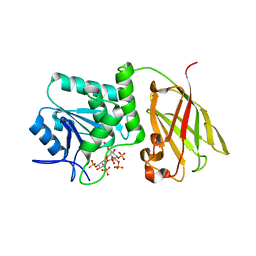 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), complexed with D-MYO-inositol-1,4,5-triphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V0F
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), form II | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2WY3
 
 | | Structure of the HCMV UL16-MICB complex elucidates select binding of a viral immunoevasin to diverse NKG2D ligands | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Mueller, S, Zocher, G, Steinle, A, Stehle, T. | | Deposit date: | 2009-11-11 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Hcmv Ul16-Micb Complex Elucidates Select Binding of a Viral Immunoevasin to Diverse Nkg2D Ligands.
Plos Pathog., 6, 2010
|
|
1KFQ
 
 | | Crystal Structure of Exocytosis-Sensitive Phosphoprotein, pp63/parafusin (Phosphoglucomutse) from Paramecium. OPEN FORM | | Descriptor: | CALCIUM ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-22 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
1KFI
 
 | | Crystal Structure of the Exocytosis-Sensitive Phosphoprotein, pp63/Parafusin (phosphoglucomutase) from Paramecium | | Descriptor: | SULFATE ION, ZINC ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-21 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
3TLP
 
 | | Crystal structure of the fourth bromodomain of human poly-bromodomain containing protein 1 (PB1) | | Descriptor: | NICKEL (II) ION, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Krojer, T, Allerston, C.K, Latwiel, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-08-30 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
4XUA
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with E11919 BAZ2-ICR analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{1-[2-(4-methyl-1H-1,2,3-triazol-1-yl)ethyl]-4-phenyl-1H-imidazol-5-yl}benzonitrile, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure Enabled Design of BAZ2-ICR, A Chemical Probe Targeting the Bromodomains of BAZ2A and BAZ2B.
J.Med.Chem., 58, 2015
|
|
4MR4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolinone ligand (RVX-208) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6XZ8
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with N-[(1R)-1-[5-(6-chloro-1,1-dimethyl-3-oxo-isoindolin-2-yl)-3-pyridyl]ethyl]methanesulfonamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, HEME C, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
2QEP
 
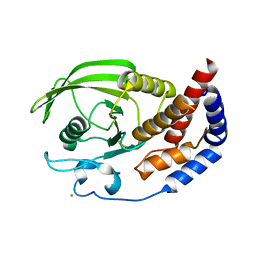 | | Crystal structure of the D1 domain of PTPRN2 (IA2beta) | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase N2 | | Authors: | Ugochukwu, E, Barr, A, Alfano, I, Berridge, G, Burgess-Brown, N, Das, S, Fedorov, O, King, O, Niesen, F, Watt, S, Savitsky, P, Salah, E, Pike, A.C.W, Bunkoczi, G, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-26 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
3BQF
 
 | |
5FE2
 
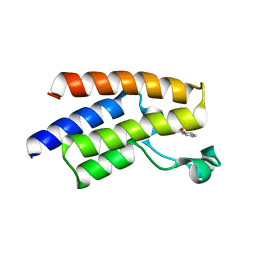 | | Crystal structure of human PCAF bromodomain in complex with fragment BR013 (fragment 3) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3~{H}-isoindol-1-one, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE9
 
 | | Crystal structure of human PCAF bromodomain in complex with compound SL1122 (compound 13) | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase KAT2B, ~{N}-(1,4-dimethyl-2-oxidanylidene-quinolin-7-yl)methanesulfonamide | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE0
 
 | | Crystal structure of human PCAF bromodomain in complex with acetyllysine | | Descriptor: | Histone acetyltransferase KAT2B, N(6)-ACETYLLYSINE | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE7
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment ZB2216 (fragment 11) | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-hydroxyethyl)-3-methyl-6,7-dihydro-5~{H}-indazol-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE6
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment ZB1916 (fragment 10) | | Descriptor: | (4-azanylpiperidin-1-yl)-cyclopropyl-methanone, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FDZ
 
 | | Crystal structure of human PCAF bromodomain in complex with compound BDOMB00091a (compound 14) | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase KAT2B, ~{N}-methyl-2-(oxan-4-yloxy)-5-(2-oxidanylidene-2-phenylazanyl-ethoxy)benzamide | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE3
 
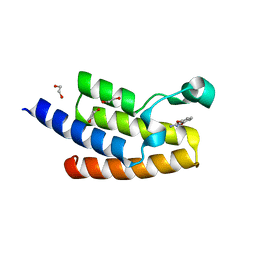 | | Crystal structure of human PCAF bromodomain in complex with fragment MB360 (fragment 4) | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1,2-benzoxazol-3-amine, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE5
 
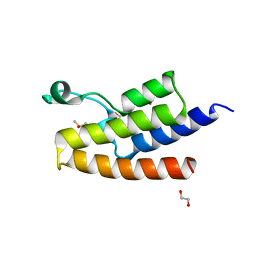 | | Crystal structure of human PCAF bromodomain in complex with fragment MB093 (fragment 7) | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(1,2,3-thiadiazol-4-yl)phenyl]methanamine, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE1
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment BR004 (fragment 1) | | Descriptor: | 1,2-ETHANEDIOL, 1-METHYLQUINOLIN-2(1H)-ONE, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE8
 
 | | Crystal structure of human PCAF bromodomain in complex with compound SL1126 (compound 12) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone acetyltransferase KAT2B, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FH7
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with compound 18 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-3-[(dimethylamino)methyl]-4~{H}-pyrrolo[1,2-a]quinazolin-5-one, Protein polybromo-1 | | Authors: | Tallant, C, Sutherell, C.L, Siejka, P, Sorrell, F.J, Krojer, T, Picaud, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Ley, S.V, Knapp, S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Identification and Development of 2,3-Dihydropyrrolo[1,2-a]quinazolin-5(1H)-one Inhibitors Targeting Bromodomains within the Switch/Sucrose Nonfermenting Complex.
J.Med.Chem., 59, 2016
|
|
