8OYX
 
 | | De novo designed soluble GPCR-like fold GLF_18 | | Descriptor: | De novo designed soluble GPCR-like protein, PHOSPHATE ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Computational design of soluble functional analogues of integral membrane proteins.
Biorxiv, 2024
|
|
8OYY
 
 | | De novo designed soluble GPCR-like fold GLF_32 | | Descriptor: | CHLORIDE ION, De novo designed soluble GPCR-like protein, POTASSIUM ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Computational design of soluble functional analogues of integral membrane proteins.
Biorxiv, 2024
|
|
8OYV
 
 | | De novo designed Claudin fold CLF_4 | | Descriptor: | De novo designed soluble Claudin | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Computational design of soluble functional analogues of integral membrane proteins.
Biorxiv, 2024
|
|
8OYS
 
 | | De novo designed TIM barrel fold TBF_24 | | Descriptor: | CHLORIDE ION, De novo designed TIM-barrel | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Computational design of soluble functional analogues of integral membrane proteins.
Biorxiv, 2024
|
|
3PPD
 
 | | GGVLVN segment from Human Prostatic Acid Phosphatase Residues 260-265, involved in Semen-Derived Enhancer of Viral Infection | | Descriptor: | ACETIC ACID, GGVLVN peptide, amyloid forming segment, ... | | Authors: | Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation.
Nature, 475, 2011
|
|
3Q6O
 
 | | Oxidoreductase Fragment of Human QSOX1 | | Descriptor: | SULFATE ION, Sulfhydryl oxidase 1 | | Authors: | Fass, D, Alon, A. | | Deposit date: | 2011-01-03 | | Release date: | 2012-05-30 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The dynamic disulphide relay of quiescin sulphydryl oxidase.
Nature, 488, 2012
|
|
3QD9
 
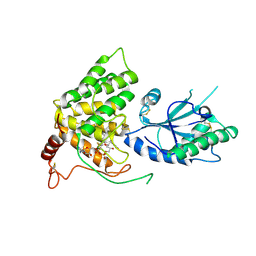 | |
4JPJ
 
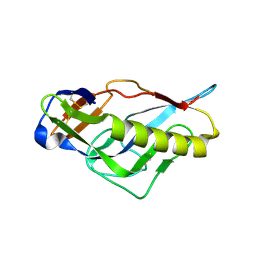 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT6 | | Authors: | Julien, J.-P, Jardine, J, Schief, W.R, Wilson, I.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational HIV immunogen design to target specific germline B cell receptors.
Science, 340, 2013
|
|
3QCP
 
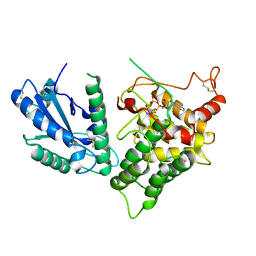 | | QSOX from Trypanosoma brucei | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QSOX from Trypanosoma brucei (TbQSOX) | | Authors: | Alon, A, Fass, D. | | Deposit date: | 2011-01-17 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The dynamic disulphide relay of quiescin sulphydryl oxidase.
Nature, 488, 2012
|
|
4JPI
 
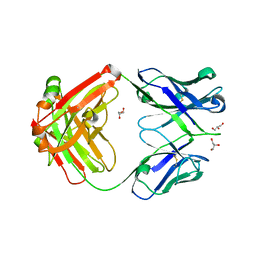 | | Crystal structure of a putative VRC01 germline precursor Fab | | Descriptor: | GLYCEROL, Putative VRC01 germline Fab heavy chain, Putative VRC01 germline Fab light chain | | Authors: | Julien, J.-P, Diwanji, D.C, Jardine, J, Schief, W.R, Wilson, I.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational HIV immunogen design to target specific germline B cell receptors.
Science, 340, 2013
|
|
4JPK
 
 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain eOD-GT6 in complex with a putative VRC01 germline precursor Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT6, ... | | Authors: | Julien, J.-P, Jardine, J, Schief, W.R, Wilson, I.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational HIV immunogen design to target specific germline B cell receptors.
Science, 340, 2013
|
|
5CY5
 
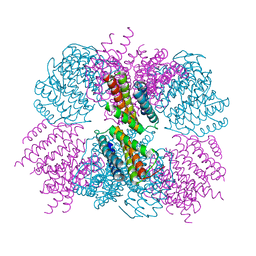 | | Crystal structure of the T33-51H designed self-assembling protein tetrahedron | | Descriptor: | T33-51H-A, T33-51H-B | | Authors: | Cannon, K.A, Cascio, D, Park, R, Boyken, S, King, N, Yeates, T.O. | | Deposit date: | 2015-07-30 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
7TYD
 
 | |
7UDK
 
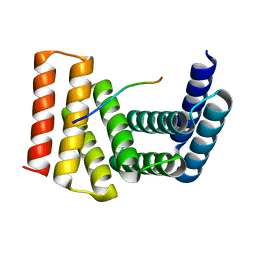 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | Descriptor: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDO
 
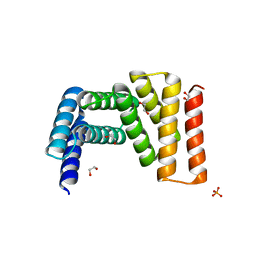 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDL
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | Descriptor: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7UDM
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 in alternative conformation 1 (with peptide) | | Descriptor: | 6xPLP, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UPQ
 
 | |
7UPP
 
 | |
7UPO
 
 | |
7UE2
 
 | |
6DG4
 
 | |
5FKA
 
 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN E, T CELL RECEPTOR ALPHA CHAIN, T CELL RECEPTOR BETA CHAIN, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two Common Structural Motifs for Tcr Recognition by Staphylococcal Enterotoxins.
Sci.Rep., 6, 2016
|
|
5FK9
 
 | |
