6Y30
 
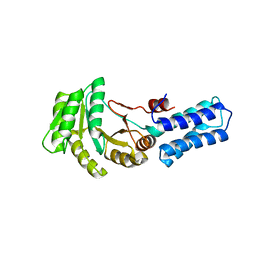 | | NG domain of human SRP54 T115A mutant | | Descriptor: | Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Wild, K, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
4WFM
 
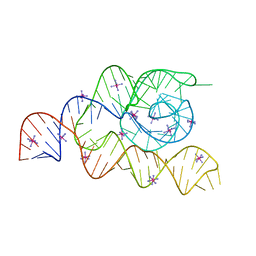 | | Structure of the complete bacterial SRP Alu domain | | Descriptor: | Bacillus subtilis small cytoplasmic RNA (scRNA),RNA, COBALT HEXAMMINE(III), MAGNESIUM ION | | Authors: | Kempf, G, Wild, K, Sinning, I. | | Deposit date: | 2014-09-15 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the complete bacterial SRP Alu domain.
Nucleic Acids Res., 42, 2014
|
|
4WFL
 
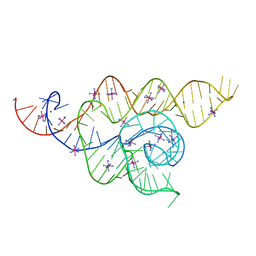 | |
4UE4
 
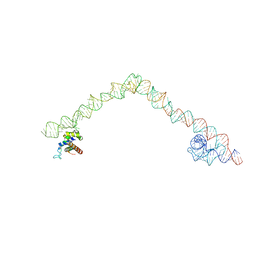 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 6S RNA, FTSQ SIGNAL SEQUENCE, SIGNAL RECOGNITION PARTICLE PROTEIN | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UE5
 
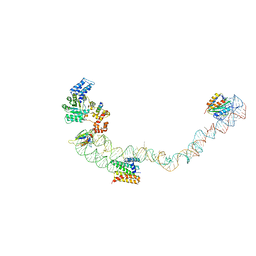 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7Q74
 
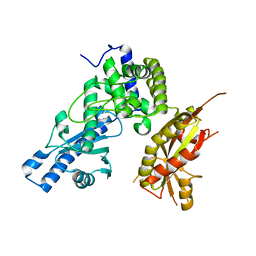 | |
7Q73
 
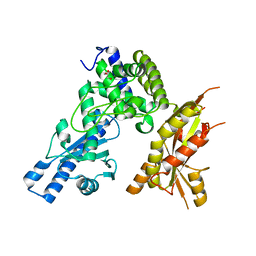 | | Structure of Pla1 apo | | Descriptor: | GLYCEROL, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
7Q72
 
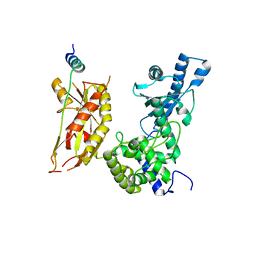 | | Structure of Pla1 in complex with Red1 | | Descriptor: | NURS complex subunit red1, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
8CR1
 
 | | Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1,GET2-GET1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
5NQH
 
 | | Structure of the human Fe65-PTB2 homodimer | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1, GLYCEROL, SULFATE ION | | Authors: | Feilen, L.P, Haubrich, K, Sinning, I, Konietzko, U, Kins, S, Simon, B, Wild, K. | | Deposit date: | 2017-04-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fe65-PTB2 Dimerization Mimics Fe65-APP Interaction.
Front Mol Neurosci, 10, 2017
|
|
5M72
 
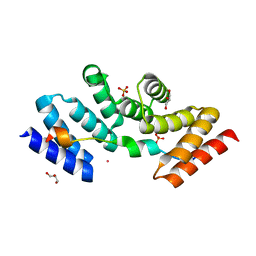 | | Structure of the human SRP68-72 protein-binding domain complex | | Descriptor: | GLYCEROL, POTASSIUM ION, SULFATE ION, ... | | Authors: | Becker, M.M.M, Wild, K, Sinning, I. | | Deposit date: | 2016-10-26 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human SRP72 complexes provide insights into SRP RNA remodeling and ribosome interaction.
Nucleic Acids Res., 45, 2017
|
|
5M73
 
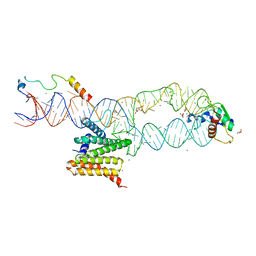 | | Structure of the human SRP S domain with SRP72 RNA-binding domain | | Descriptor: | GLYCEROL, Human gene for small cytoplasmic 7SL RNA (7L30.1), MAGNESIUM ION, ... | | Authors: | Becker, M.M.M, Wild, K, Sinning, I. | | Deposit date: | 2016-10-26 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human SRP72 complexes provide insights into SRP RNA remodeling and ribosome interaction.
Nucleic Acids Res., 45, 2017
|
|
8CR2
 
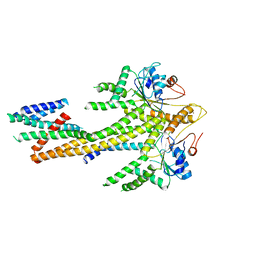 | | Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8CQZ
 
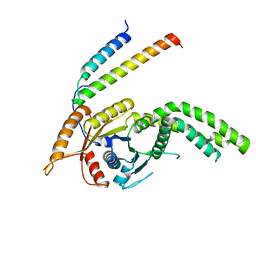 | | Homo sapiens Get3 in complex with the Get1 cytoplasmic domain | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor 1 | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Saar, D, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
3IQX
 
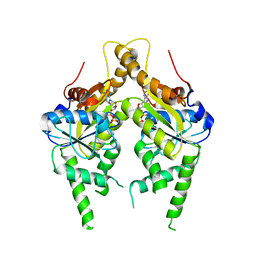 | | ADP complex of C.therm. Get3 in closed form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IQW
 
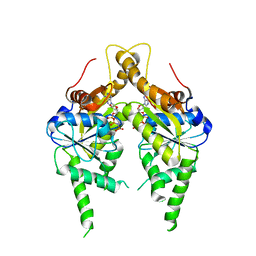 | | AMPPNP complex of C. therm. Get3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6RY3
 
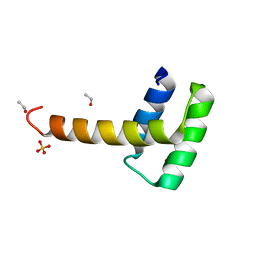 | | Structure of the WUS homeodomain | | Descriptor: | ACETYL GROUP, Protein WUSCHEL, SULFATE ION | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6SO5
 
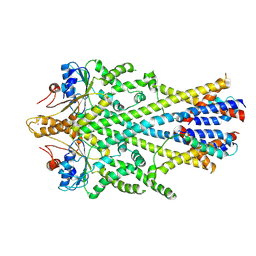 | | Homo sapiens WRB/CAML heterotetramer in complex with a TRC40 dimer | | Descriptor: | ATPase ASNA1, Calcium signal-modulating cyclophilin ligand, Tail-anchored protein insertion receptor WRB, ... | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Flemming, D, Sinning, I. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-09 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex.
Mol.Cell, 80, 2020
|
|
8ODV
 
 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (nanodisc) | | Descriptor: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | Authors: | McDowell, M.A, Wild, K, Sinning, I. | | Deposit date: | 2023-03-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8ODU
 
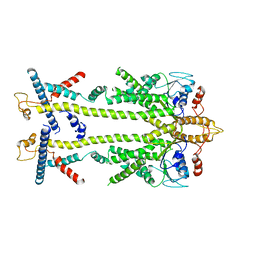 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (amphipol) | | Descriptor: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | Authors: | McDowell, M.A, Wild, K, Sinning, I. | | Deposit date: | 2023-03-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8OO0
 
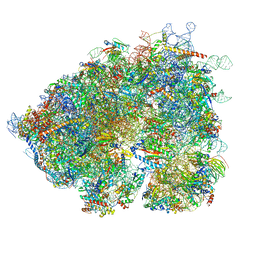 | | Chaetomium thermophilum Methionine Aminopeptidase 2 autoproteolysis product at the 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S0, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
6RYI
 
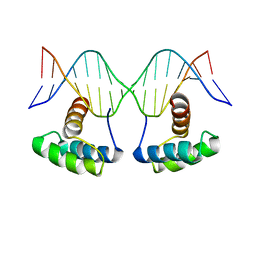 | | WUS-HD bound to G-Box DNA | | Descriptor: | DNA (5'-D(P*CP*CP*CP*AP*TP*CP*AP*CP*GP*TP*GP*AP*CP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*TP*CP*AP*CP*GP*TP*GP*AP*TP*GP*GP*G)-3'), Protein WUSCHEL | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6RYD
 
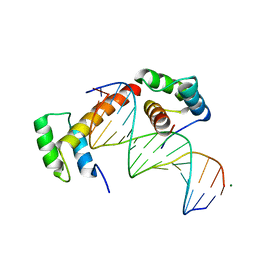 | | WUS-HD bound to TGAA DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*GP*TP*AP*TP*GP*AP*AP*TP*GP*AP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*TP*CP*AP*TP*TP*CP*AP*TP*AP*CP*AP*CP*T)-3'), MAGNESIUM ION, ... | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.575 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6RYL
 
 | | WUS-HD bound to TAAT DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*AP*AP*CP*CP*CP*AP*TP*TP*AP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*TP*AP*AP*TP*GP*GP*GP*TP*TP*GP*TP*G)-3'), Protein WUSCHEL | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
8ONX
 
 | | High resolution structure of Chaetomium thermophilum MAP2 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
