7ET8
 
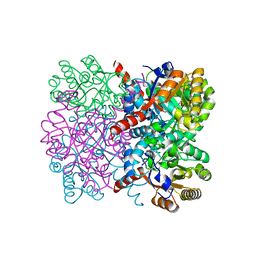 | | Crystal structure of thermostable AbHpaI, a class II metal dependent pyruvate aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETI
 
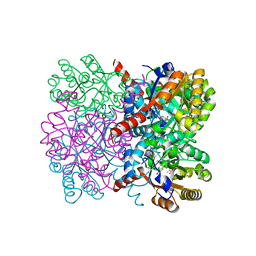 | | Crystal structure of AbHpaI-Zn-pyruvate-4-hydroxybenzaldehyde complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, P-HYDROXYBENZALDEHYDE, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETD
 
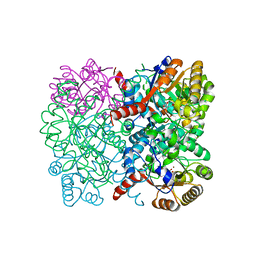 | | Crystal structure of AbHpaI-Zn-(4S)-KDGlu complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETC
 
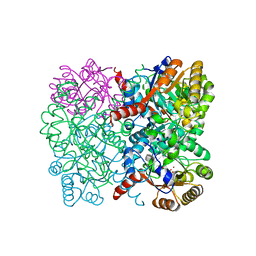 | | Crystal structure of AbHpaI-Zn-(4R)-KDGal complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | (4R,5R)-4,5,6-tris(oxidanyl)-2-oxidanylidene-hexanoic acid, 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETE
 
 | | Crystal structure of AbHpaI-Mg-(4R)-KDGal complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | (4R,5R)-4,5,6-tris(oxidanyl)-2-oxidanylidene-hexanoic acid, 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETG
 
 | | Crystal structure of AbHpaI-Co-pyruvate-succinic semialdehyde complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, 4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETA
 
 | | Crystal structure of AbHpaI-Co-pyruvate complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETH
 
 | | Crystal structure of AbHpaI-Zn-pyruvate-propionaldehyde complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, PROPANAL, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETF
 
 | | Crystal structure of AbHpaI-Mn-pyruvate-succinic semialdehyde complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, 4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ET9
 
 | | Crystal structure of AbHpaI-Zn-pyruvate complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, PYRUVIC ACID, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
6A2P
 
 | | Crystal structure of quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum DHFR-TS complexed with BT3, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5,5'-[propane-1,3-diylbis(oxy-4,1-phenylene)]bis(6-ethylpyrimidine-2,4-diamine), Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Tarnchampoo, B, Yuthavong, Y. | | Deposit date: | 2018-06-12 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
6A2O
 
 | | Crystal structure of wild type Plasmodium falciparum DHFR-TS complexed with BT3, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5,5'-[propane-1,3-diylbis(oxy-4,1-phenylene)]bis(6-ethylpyrimidine-2,4-diamine), Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Tarnchampoo, B, Yuthavong, Y. | | Deposit date: | 2018-06-12 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
6A2K
 
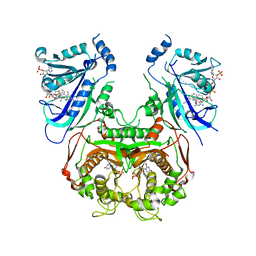 | | Crystal structure of wild type Plasmodium falciparum DHFR-TS complexed with BT1, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(3-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)-6-ethylpyrimidine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Tarnchampoo, B, Yuthavong, Y. | | Deposit date: | 2018-06-12 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
6A2M
 
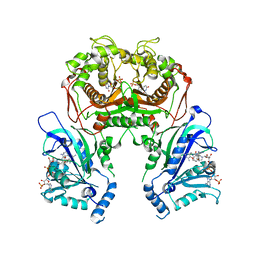 | | Crystal structure of wild type Plasmodium falciparum DHFR-TS complexed with BT2, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)-6-ethylpyrimidine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Tarnchampoo, B, Yuthavong, Y. | | Deposit date: | 2018-06-12 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
7ETB
 
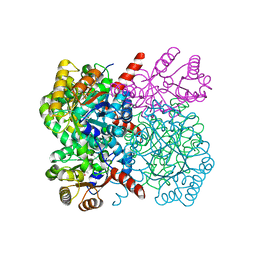 | | Crystal structure of AbHpaI-Mn-pyruvate complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
4O6Z
 
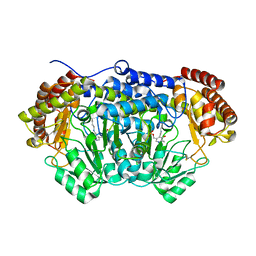 | | Crystal structure of serine hydroxymethyltransferase with covalently bound PLP Schiff-base from Plasmodium falciparum | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2013-12-24 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The structure of Plasmodium falciparum serine hydroxymethyltransferase reveals a novel redox switch that regulates its activities.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OYT
 
 | | Crystal structure of ternary complex of Plasmodium vivax SHMT with D-serine and folinic acid | | Descriptor: | (2R)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-oxidanyl-propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2014-02-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PFN
 
 | | Crystal structure of Plasmodium vivax SHMT with L-serine Schiff base | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, SERINE, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2014-04-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PFF
 
 | | Crystal structure of Plasmodium vivax SHMT with PLP Schiff base | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, putative | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2014-04-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5YG0
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS657 | | Descriptor: | 2-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]piperidin-4-yl]ethanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | to be published
to be published
|
|
5YFY
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS625 | | Descriptor: | 1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-fluoranyl-phenyl]piperidine-4-carboxylic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | to be published
to be published
|
|
5YG1
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS704 | | Descriptor: | 3-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]piperidin-4-yl]propanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | to be published
to be published
|
|
7E8P
 
 | | Crystal structure of a Flavin-dependent Monooxygenase HadA wild type complexed with reduced FAD and 4-nitrophenol | | Descriptor: | Chlorophenol monooxygenase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, P-NITROPHENOL | | Authors: | Pimviriyakul, P, Jaruwat, A, Chitnumsub, P, Chaiyen, P. | | Deposit date: | 2021-03-02 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into a flavin-dependent dehalogenase HadA explain catalysis and substrate inhibition via quadruple pi-stacking.
J.Biol.Chem., 297, 2021
|
|
7E8Q
 
 | | Crystal structure of a Flavin-dependent Monooxygenase HadA F441V mutant complexed with reduced FAD and 4-nitrophenol | | Descriptor: | Chlorophenol monooxygenase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, P-NITROPHENOL | | Authors: | Pimviriyakul, P, Jaruwat, A, Chitnumsub, P, Chaiyen, P. | | Deposit date: | 2021-03-02 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into a flavin-dependent dehalogenase HadA explain catalysis and substrate inhibition via quadruple pi-stacking.
J.Biol.Chem., 297, 2021
|
|
5GYH
 
 | |
