8TA4
 
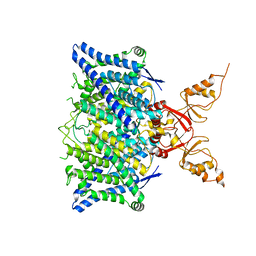 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA5
 
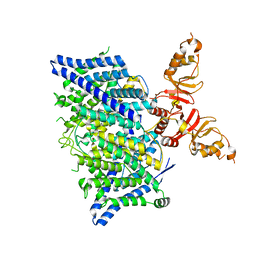 | | Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA2
 
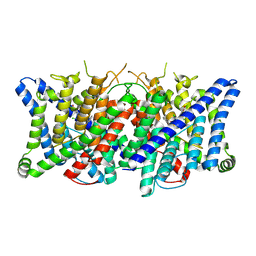 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42 | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA6
 
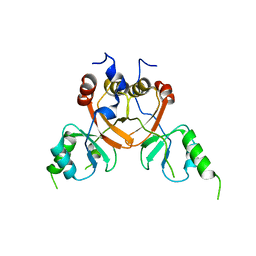 | | Cryo-EM structure of the human CLC-2 chloride channel C-terminal domain | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA3
 
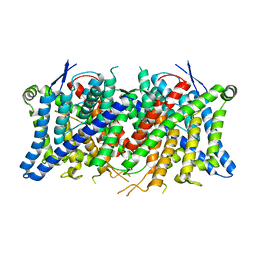 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain Apo state with resolved N-terminal hairpin | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8FR7
 
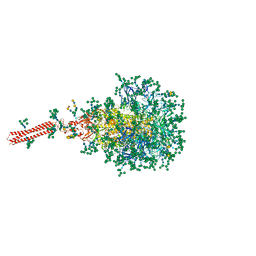 | | A hinge glycan regulates spike bending and impacts coronavirus infectivity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pintilie, G, Wilson, E, Chmielewski, D, Schmid, M.F, Jin, J, Chen, M, Singharoy, A, Chiu, W. | | Deposit date: | 2023-01-06 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A hinge glycan regulates spike bending and impacts coronavirus infectivity
To Be Published
|
|
8FCG
 
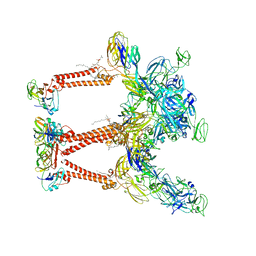 | | Cryo-EM structure of Chikungunya virus asymmetric unit | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Capsid protein, E1 glycoprotein, ... | | Authors: | Su, G.C, Chmielewsk, D, Kaelber, J, Pintilie, G, Chen, M, Jin, J, Auguste, A, Chiu, W. | | Deposit date: | 2022-12-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryogenic electron microscopy and tomography reveal imperfect icosahedral symmetry in alphaviruses.
Pnas Nexus, 3, 2024
|
|
5UU5
 
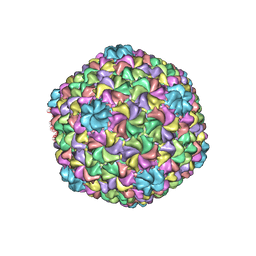 | | Bacteriophage P22 mature virion capsid protein | | Descriptor: | Major capsid protein | | Authors: | Hryc, C.F, Chen, D.-H, Afonine, P.V, Jakana, J, Wang, Z, Haase-Pettingell, C, Jiang, W, Adams, P.D, King, J.A, Schmid, M.F, Chiu, W. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Accurate model annotation of a near-atomic resolution cryo-EM map.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7R9J
 
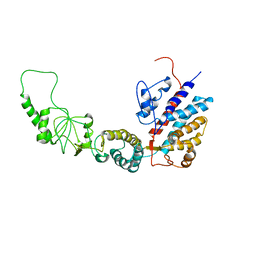 | | Methanococcus maripaludis chaperonin, open conformation 4 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9I
 
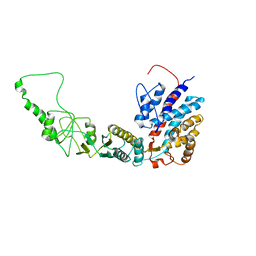 | | Methanococcus maripaludis chaperonin, open conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9K
 
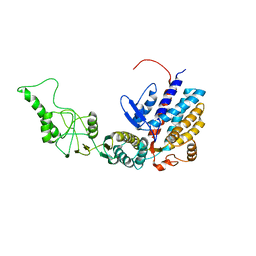 | | Methanococcus maripaludis chaperonin, closed conformation 4 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9H
 
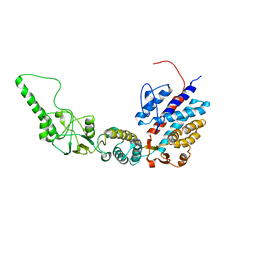 | | Methanococcus maripaludis chaperonin, open conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9M
 
 | | Methanococcus maripaludis chaperonin, closed conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9E
 
 | | Methanococcus maripaludis chaperonin, open conformation 1 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7RAK
 
 | | Methanococcus maripaludis chaperonin complex in open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9O
 
 | | Methanococcus maripaludis chaperonin, closed conformation 1 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9U
 
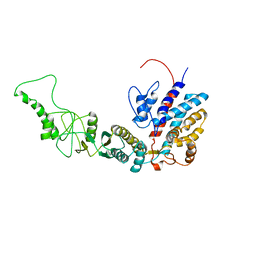 | | Methanococcus maripaludis chaperonin, closed conformation 3 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
6C6L
 
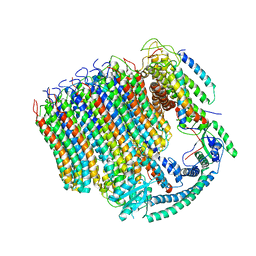 | | Yeast Vacuolar ATPase Vo in lipid nanodisc | | Descriptor: | V-type proton ATPase subunit a, vacuolar isoform, V-type proton ATPase subunit c, ... | | Authors: | Roh, S, Stam, N.J, Hryc, C, Couoh-Cardel, S, Pintilie, G, Chiu, W, Wilkens, S. | | Deposit date: | 2018-01-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The 3.5- angstrom CryoEM Structure of Nanodisc-Reconstituted Yeast Vacuolar ATPase VoProton Channel.
Mol. Cell, 69, 2018
|
|
7TBH
 
 | | cryo-EM structure of MBP-KIX-apoferritin complex with peptide 7 | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed, LEU-SER-ARG-ARG-PRO-SEP-TYR-ARG-LYS-ILE-LEU-ASN-ASP-LEU-SER-SER-ASP-ALA-PRO | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
7TB3
 
 | | cryo-EM structure of MBP-KIX-apoferritin | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
7TNS
 
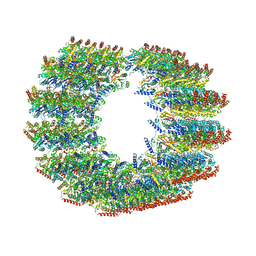 | | Subpellicular microtubule from detergent-extract Toxoplasma gondii cells | | Descriptor: | Microtubule associated protein SPM1, PDI family protein, Tubulin alpha chain, ... | | Authors: | Sun, S.Y, Pintilie, G.D, Chen, M, Chiu, W. | | Deposit date: | 2022-01-21 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo-ET of Toxoplasma parasites gives subnanometer insight into tubulin-based structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TNT
 
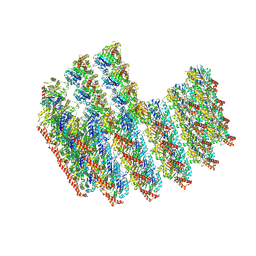 | | The tubulin-based conoid from detergent-extract Toxoplasma gondii cells | | Descriptor: | Tubulin alpha chain, Tubulin beta chain | | Authors: | Sun, S.Y, Pintilie, G.D, Chen, M, Chiu, W. | | Deposit date: | 2022-01-21 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-ET of Toxoplasma parasites gives subnanometer insight into tubulin-based structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7JM3
 
 | | Full-length three-dimensional structure of the influenza A virus M1 protein and its organization into a matrix layer | | Descriptor: | Matrix protein 1 | | Authors: | Su, Z, Pintilie, G, Selzer, L, Chiu, W, Kirkegaard, K. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Full-length three-dimensional structure of the influenza A virus M1 protein and its organization into a matrix layer.
Plos Biol., 18, 2020
|
|
7TTN
 
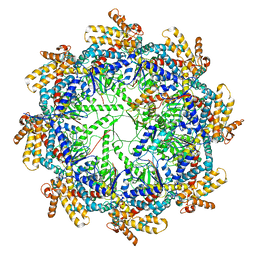 | | The beta-tubulin folding intermediate II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TRG
 
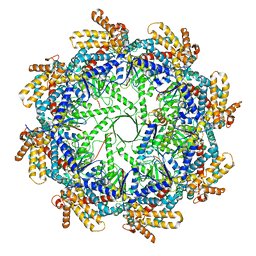 | | The beta-tubulin folding intermediate I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
