4GYC
 
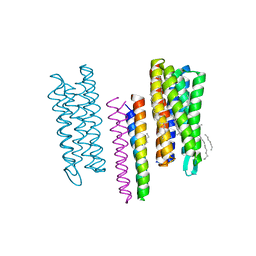 | | Structure of the SRII(D75N mutant)/HtrII Complex in I212121 space group ("U" shape) | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2012-09-05 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0501 Å) | | Cite: | Ground state structure of D75N mutant of sensory rhodopsin II in complex with its cognate transducer.
J Photochem Photobiol B, 123C, 2013
|
|
6EIG
 
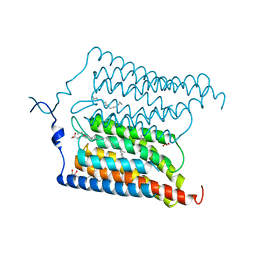 | | Crystal structure of N24Q/C128T mutant of Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, EICOSANE, ... | | Authors: | Kovalev, K, Borshchevskiy, V, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
3QDC
 
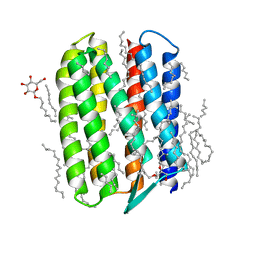 | | Crystal structure of Natronomonas pharaonis sensory rhodopsin II in the active state | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, EICOSANE, RETINAL, ... | | Authors: | Gushchin, I, Reshetnyak, A, Borshchevskiy, V, Ishchenko, A, Round, E, Grudinin, S, Engelhard, M, Buldt, G, Gordeliy, V. | | Deposit date: | 2011-01-18 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active State of Sensory Rhodopsin II: Structural Determinants for Signal Transfer and Proton Pumping.
J.Mol.Biol., 412, 2011
|
|
3QAP
 
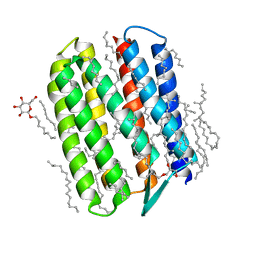 | | Crystal structure of Natronomonas pharaonis sensory rhodopsin II in the ground state | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, EICOSANE, RETINAL, ... | | Authors: | Gushchin, I, Reshetnyak, A, Borshchevskiy, V, Ishchenko, A, Round, E, Grudinin, S, Engelhard, M, Buldt, G, Gordeliy, V. | | Deposit date: | 2011-01-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active State of Sensory Rhodopsin II: Structural Determinants for Signal Transfer and Proton Pumping.
J.Mol.Biol., 412, 2011
|
|
5JTB
 
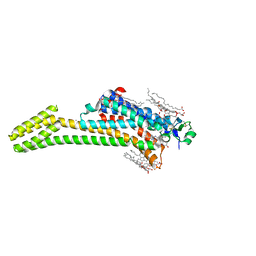 | | Crystal structure of the chimeric protein of A2aAR-BRIL with bound iodide ions | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Melnikov, I, Polovinkin, V, Shevtsov, M, Borshchevskiy, V, Cherezov, V, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
4MND
 
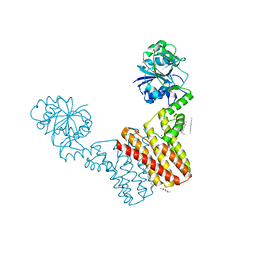 | | Crystal structure of Archaeoglobus fulgidus IPCT-DIPPS bifunctional membrane protein | | Descriptor: | CTP L-myo-inositol-1-phosphate cytidylyltransferase/CDP-L-myo-inositol myo-inositolphosphotransferase, EICOSANE, MAGNESIUM ION | | Authors: | Nogly, P, Gushchin, I, Remeeva, A, Esteves, A.M, Ishchenko, A, Ma, P, Grudinin, S, Borges, N, Round, E, Moraes, I, Borshchevskiy, V, Santos, H, Gordeliy, V, Archer, M. | | Deposit date: | 2013-09-10 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | X-ray structure of a CDP-alcohol phosphatidyltransferase membrane enzyme and insights into its catalytic mechanism.
Nat Commun, 5, 2014
|
|
5JJN
 
 | | Structure of the SRII/HtrII(G83F) Complex in P212121 space group ("V" shape) | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2016-04-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
5JJE
 
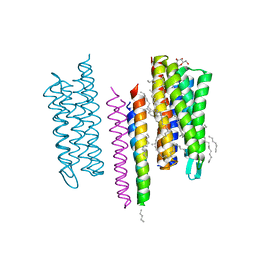 | | Structure of the SRII/HtrII Complex in I212121 space group ("U" shape) | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2016-04-23 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
8RT2
 
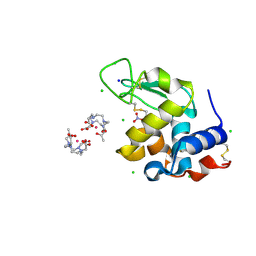 | | Hen Egg White Lysozyme Crystallized with Bioassembler on Earth | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | MacCarthy, C, Koudan, E.V, Petrov, S.V, Levin, A.A, Khesuani, Y.D, Borshchevskiy, V. | | Deposit date: | 2024-01-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Hen Egg White Lysozyme Crystallized with Bioassembler on Earth
To Be Published
|
|
8RT3
 
 | | Hen Egg White Lysozyme Crystallized with Bioassembler in Space Microgravity Conditions | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | MacCarthy, C, Koudan, E.V, Petrov, S.V, Levin, A.A, Khesuani, Y.D, Borshchevskiy, V. | | Deposit date: | 2024-01-25 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Hen Egg White Lysozyme Crystallized with Bioassembler in Space Microgravity Conditions
To Be Published
|
|
8BBW
 
 | | Crystal structure of medical leech destabilase (low salt) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme | | Authors: | Marin, E, Bukhdruker, S, Manuvera, V, Kornilov, D, Zinovev, E, Bobrovsky, P, Lazarev, V, Borshchevskiy, V. | | Deposit date: | 2022-10-14 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for inhibition of thrombolytic destabilase
from medical leech by physiological sodium concentrations
To Be Published
|
|
6RZ5
 
 | | XFEL crystal structure of the human cysteinyl leukotriene receptor 1 in complex with zafirlukast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Cysteinyl leukotriene receptor 1,Soluble cytochrome b562,Cysteinyl leukotriene receptor 1, OLEIC ACID, ... | | Authors: | Luginina, A, Gusach, A, Marin, E, Mishin, A, Brouillette, R, Popov, P, Shiryaeva, A, Besserer-Offroy, E, Longpre, J.M, Lyapina, E, Ishchenko, A, Patel, N, Polovinkin, V, Safronova, N, Bogorodskiy, A, Edelweiss, E, Liu, W, Batyuk, A, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs.
Sci Adv, 5, 2019
|
|
7A9A
 
 | | Crystal structure of rubredoxin B (Rv3250c) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vakhrameev, D, Kavaleuski, A, Bukhdruker, S, Marin, E, Sushko, T, Grabovec, I.P, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2020-09-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A new twist of rubredoxin function in M. tuberculosis.
Bioorg.Chem., 109, 2021
|
|
8AMO
 
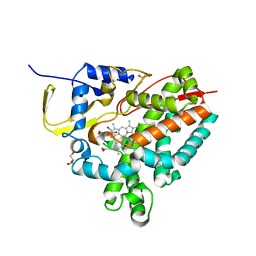 | | Crystal structure of M. tuberculosis CYP143 | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Grudo, A, Marin, E, Kapranov, I, Shevtsov, M, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMP
 
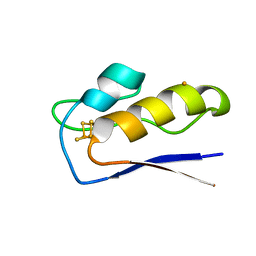 | | Crystal structure of M.tuberculosis ferredoxin Fdx | | Descriptor: | FE (III) ION, FE3-S4 CLUSTER, Possible ferredoxin | | Authors: | Bukhdruker, S, Kavaleuski, A, Marin, E, Kapranov, I, Mishin, A, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMQ
 
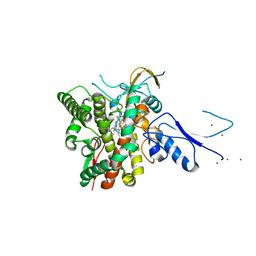 | | Crystal structure of the complex CYP143-FdxE from M. tuberculosis | | Descriptor: | FE3-S4 CLUSTER, NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Smolskaya, S, Marin, E, Kapranov, I, Kovalev, K, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
7AV6
 
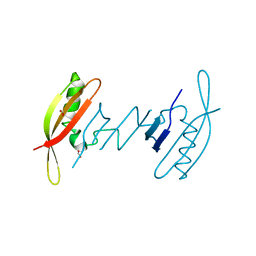 | | FAST in a domain-swapped dimer form | | Descriptor: | FORMIC ACID, Photoactive yellow protein | | Authors: | Bukhdruker, S, Remeeva, A, Ruchkin, D, Gorbachev, D, Povarova, N, Mineev, K, Goncharuk, S, Baranov, M, Mishin, A, Borshchevskiy, V. | | Deposit date: | 2020-11-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NanoFAST: structure-based design of a small fluorogen-activating protein with only 98 amino acids.
Chem Sci, 12, 2021
|
|
6RZ4
 
 | | Crystal structure of cysteinyl leukotriene receptor 1 in complex with pranlukast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Cysteinyl leukotriene receptor 1,Soluble cytochrome b562,Cysteinyl leukotriene receptor 1, OLEIC ACID, ... | | Authors: | Luginina, A, Gusach, A, Marin, E, Mishin, A, Brouillette, R, Popov, P, Shiryaeva, A, Besserer-Offroy, E, Longpre, J.M, Lyapina, E, Ishchenko, A, Patel, N, Polovinkin, V, Safronova, N, Bogorodskiy, A, Edelweiss, E, Liu, W, Batyuk, A, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs.
Sci Adv, 5, 2019
|
|
6RF1
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric "wet" form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF9
 
 | | Crystal structure of the Y154F mutant of the light-driven sodium pump KR2 in the monomeric form, pH 8.0 | | Descriptor: | EICOSANE, GLYCEROL, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RFC
 
 | | Crystal structure of the potassium-pumping G263F mutant of the light-driven sodium pump KR2 in the monomeric form, pH 4.3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF0
 
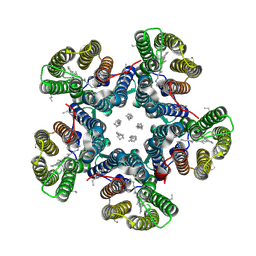 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric "dry" form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6REX
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form, pH 6.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF4
 
 | | Crystal structure of the potassium-pumping S254A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF3
 
 | | Crystal structure of the potassium-pumping G263F mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, SODIUM ION, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
