2E3Z
 
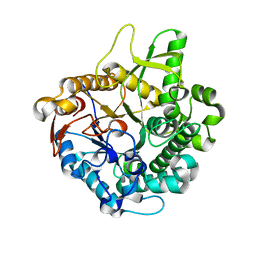 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in substrate-free form | | Descriptor: | Beta-glucosidase | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
2EGY
 
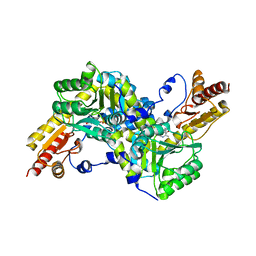 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (substrate free form), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2007-03-02 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of LysN, alpha-aminoadipate aminotransferase, from Thermus thermophilus HB27
To be Published
|
|
2E2P
 
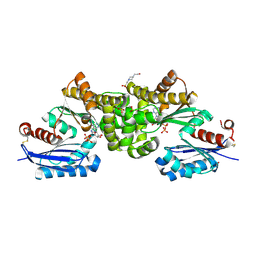 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
2E2Q
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with xylose, Mg2+, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE, MAGNESIUM ION, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
3ACF
 
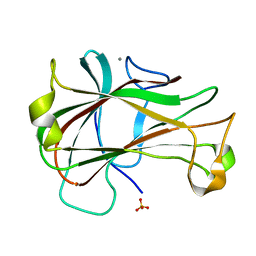 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in a ligand-free form | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, SULFATE ION | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
2E2N
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in the apo form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEXOKINASE, SULFATE ION | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
3ACG
 
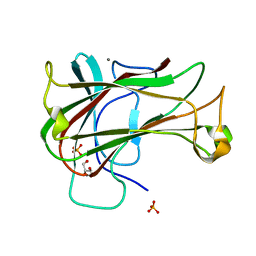 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellobiose | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, GLYCEROL, ... | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACH
 
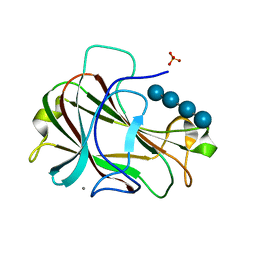 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellotetraose | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACI
 
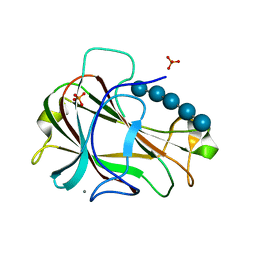 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellopentaose | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
2E40
 
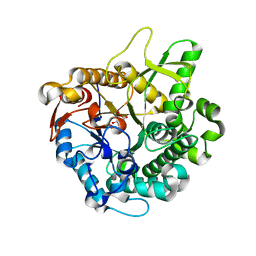 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in complex with gluconolactone | | Descriptor: | Beta-glucosidase, D-glucono-1,5-lactone | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
2E2O
 
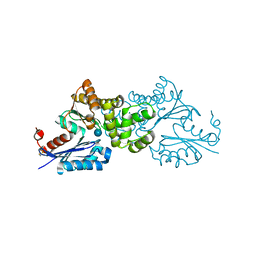 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with glucose | | Descriptor: | HEXOKINASE, beta-D-glucopyranose | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
1UGS
 
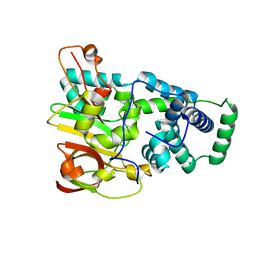 | | Crystal structure of aY114T mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGR
 
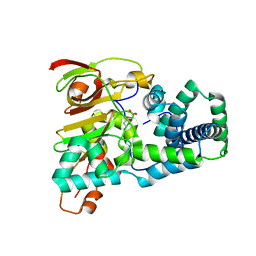 | | Crystal structure of aT109S mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGQ
 
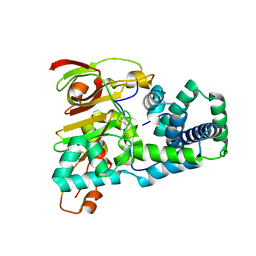 | | Crystal structure of apoenzyme of Co-type nitrile hydratase | | Descriptor: | Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1V9T
 
 | | Structure of E. coli cyclophilin B K163T mutant bound to succinyl-ALA-PRO-ALA-P-nitroanilide | | Descriptor: | (SIN)APA(NIT), cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2004-02-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
1ULW
 
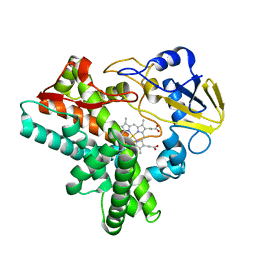 | | Crystal structure of P450nor Ser73Gly/Ser75Gly mutant | | Descriptor: | Cytochrome P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Su, F, Li, Z, Takaya, N, Shoun, H. | | Deposit date: | 2003-09-16 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
1UMG
 
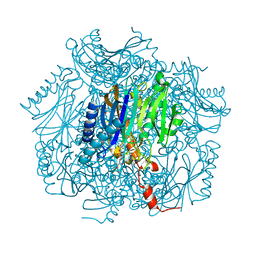 | | Crystal structure of fructose-1,6-bisphosphatase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), 385aa long conserved hypothetical protein, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2003-09-30 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea.
Structure, 12, 2004
|
|
1VAI
 
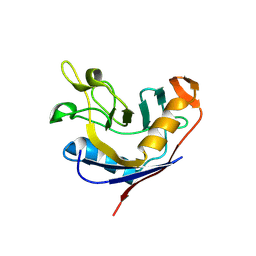 | | Structure of e. coli cyclophilin B K163T mutant bound to n-acetyl-ala-ala-pro-ala-7-amino-4-methylcoumarin | | Descriptor: | (ACE)AAPA(MCM), cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2004-02-17 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
1V94
 
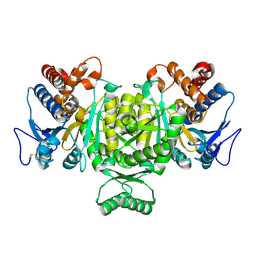 | | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix | | Descriptor: | isocitrate dehydrogenase | | Authors: | Jeong, J.-J, Sonoda, T, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2004-01-20 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix
Proteins, 55, 2004
|
|
1VCK
 
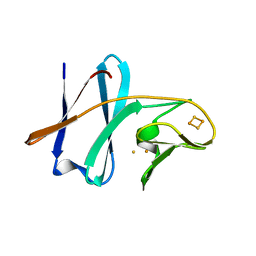 | | Crystal structure of ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10 | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, HYDROSULFURIC ACID, ... | | Authors: | Nam, J.-W, Noguchi, H, Fujiomoto, Z, Mizuno, H, Fushinobu, S, Kobashi, N, Iwata, K, Yoshida, T, Habe, H, Yamane, H, Omori, T, Nojiri, H. | | Deposit date: | 2004-03-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10, a novel Rieske non-heme iron oxygenase system
PROTEINS, 58, 2005
|
|
1WD3
 
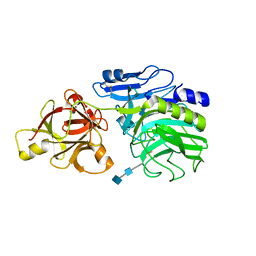 | | Crystal structure of arabinofuranosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2004-05-11 | | Release date: | 2004-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose
J.Biol.Chem., 279, 2004
|
|
1X0L
 
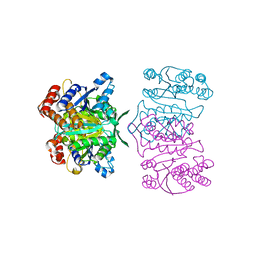 | | Crystal structure of tetrameric homoisocitrate dehydrogenase from an extreme thermophile, Thermus thermophilus | | Descriptor: | Homoisocitrate dehydrogenase | | Authors: | Miyazaki, J, Asada, K, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2005-03-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Tetrameric Homoisocitrate Dehydrogenase from an Extreme Thermophile, Thermus thermophilus: Involvement of Hydrophobic Dimer-Dimer Interaction in Extremely High Thermotolerance
J.Bacteriol., 187, 2005
|
|
1WD4
 
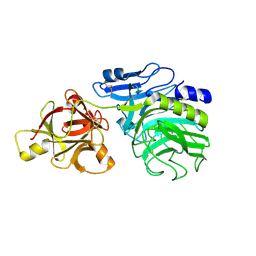 | | Crystal structure of arabinofuranosidase complexed with arabinose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2004-05-11 | | Release date: | 2004-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose
J.Biol.Chem., 279, 2004
|
|
1XQD
 
 | | Crystal structure of P450NOR complexed with 3-pyridinealdehyde adenine dinucleotide | | Descriptor: | CYTOCHROME P450 55A1, NICOTINIC ACID ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Takaya, N, Su, F, Wakagi, T, Shoun, H. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
3UES
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
