7RJF
 
 | | MOPD-1 mutant-L47W | | Descriptor: | MALONATE ION, ZINC ION, [L47W]MOPD-1 | | Authors: | Huawu, Y, Conan, K.W, Gordon, J.K, Brett, M.C, Yen-Hua, H, David, J.C. | | Deposit date: | 2021-07-20 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of Potent Peptide Inhibitors of the PD-1:PD-L1 Interaction for Cancer Immunotherapy.
J.Am.Chem.Soc., 143, 2021
|
|
4E0R
 
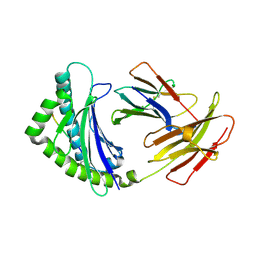 | | Structure of the chicken MHC class I molecule BF2*0401 | | Descriptor: | 8-MERIC PEPTIDE (FUS/TLS), Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
8IG4
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with GC376 | | Descriptor: | Non-structural protein 11, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
3NTZ
 
 | | Design, Synthesis, Biological Evaluation and X-ray Crystal Structures of Novel Classical 6,5,6-tricyclicbenzo[4,5]thieno[2,3-d]pyrimidines as Dual Thymidylate Synthase and Dihydrofolate Reductase Inhibitors | | Descriptor: | Dihydrofolate reductase, N-[(4-{[(2-amino-4-oxo-3,4-dihydro[1]benzothieno[2,3-d]pyrimidin-5-yl)methyl]amino}phenyl)carbonyl]-L-glutamic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-06 | | Release date: | 2011-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray crystal structure of novel classical 6,5,6-tricyclic benzo[4,5]thieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 19, 2011
|
|
3NU0
 
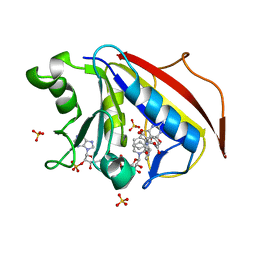 | | Design, Synthesis, Biological Evaluation and X-ray Crystal Structure of Novel Classical 6,5,6-TricyclicBenzo[4,5]thieno[2,3-d]pyrimidines as Dual Thymidylate Synthase and Dihydrofolate Reductase Inhibitors | | Descriptor: | (2S)-2-(5-{[(2-amino-4-oxo-3,4-dihydro[1]benzothieno[2,3-d]pyrimidin-5-yl)methyl]amino}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)pentanedioic acid, Dihydrofolate reducatase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-06 | | Release date: | 2011-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray crystal structure of novel classical 6,5,6-tricyclic benzo[4,5]thieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 19, 2011
|
|
7JG3
 
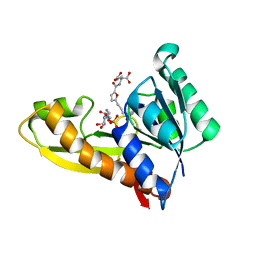 | | Human GAR transformylase in complex with GAR substrate and AGF103 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]furan-2-carbonyl}-L-glutamic acid, SODIUM ION, ... | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2020-07-18 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
7JG4
 
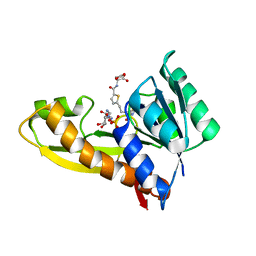 | | Human GAR transformylase in complex with GAR substrate and AGF131 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-(5-{3-[(1S,7R,8R,9S)-4-amino-2-oxo-7lambda~4~-thia-3,5-diazatetracyclo[4.3.0.0~1,7~.0~7,9~]nona-3,5-dien-8-yl]propyl}thiophene-2-carbonyl)-L-glutamic acid, SODIUM ION, ... | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2020-07-18 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
7JG0
 
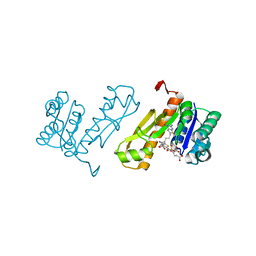 | | Human GAR transformylase in complex with GAR substrate and AGF102 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]thiophene-2-carbonyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2020-07-18 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
5AIS
 
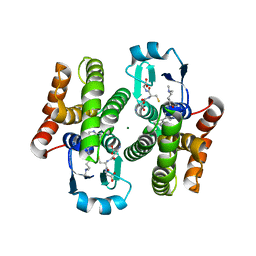 | | Complex of human hematopoietic prostagandin D2 synthase (hH-PGDS) in complex with an active site inhibitor. | | Descriptor: | 4-(dimethylamino)-N-[5-(1H-indol-4-yl)pyridin-3-yl]butanamide, GLUTATHIONE, HEMATOPOIETIC PROSTAGLANDIN D SYNTHASE, ... | | Authors: | Edfeldt, F, Evenas, J, Lepisto, M, Ward, A, Petersen, J, Wissler, L, Rohman, M, Sivars, U, Svensson, K, Perry, M, Feierberg, I, Zhou, X, Hansson, T, Narjes, F. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of Indole Inhibitors of Human Hematopoietic Prostaglandin D2 Synthase (Hh-Pgds).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5AIX
 
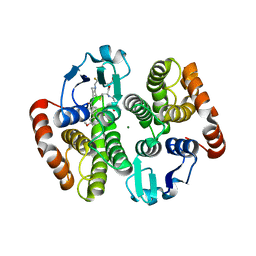 | | Complex of human hematopoietic prostagandin D2 synthase (hH-PGDS) in complex with an active site inhibitor. | | Descriptor: | 6-(3-methoxyphenyl)-N-[1-(2,2,2-trifluoroethyl)piperidin-4-yl]pyridine-3-carboxamide, GLUTATHIONE, HEMATOPOIETIC PROSTAGLANDIN D SYNTHASE, ... | | Authors: | Edfeldt, F, Evenas, J, Lepisto, M, Ward, A, Petersen, J, Wissler, L, Rohman, M, Sivars, U, Svensson, K, Perry, M, Feierberg, I, Zhou, X, Hansson, T, Narjes, F. | | Deposit date: | 2015-02-18 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Indole Inhibitors of Human Hematopoietic Prostaglandin D2 Synthase (Hh-Pgds).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5AIV
 
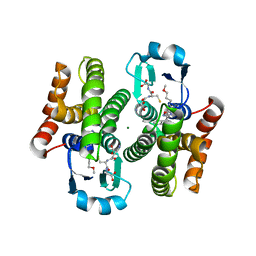 | | Complex of human hematopoietic prostagandin D2 synthase (hH-PGDS) in complex with an active site inhibitor. | | Descriptor: | 3-(1H-indol-4-yl)-N-(3-methoxypropyl)-1,2,4-oxadiazole-5-carboxamide, GLUTATHIONE, HEMATOPOIETIC PROSTAGLANDIN D SYNTHASE, ... | | Authors: | Edfeldt, F, Evenas, J, Lepisto, M, Ward, A, Petersen, J, Wissler, L, Rohman, M, Sivars, U, Svensson, K, Perry, M, Feierberg, I, Zhou, X, Hansson, T, Narjes, F. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Identification of Indole Inhibitors of Human Hematopoietic Prostaglandin D2 Synthase (Hh-Pgds).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8HCI
 
 | |
8HI8
 
 | |
8HI7
 
 | |
5YGI
 
 | |
2JV0
 
 | | SET domain of RIZ1 tumor suppressor (PRDM2) | | Descriptor: | PR domain zinc finger protein 2 | | Authors: | Briknarova, K. | | Deposit date: | 2007-09-10 | | Release date: | 2008-01-22 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the SET domain from RIZ1 tumor suppressor
Biochem.Biophys.Res.Commun., 366, 2008
|
|
8HVZ
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8UPV
 
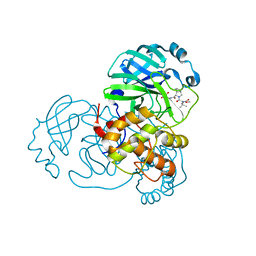 | | Structure of SARS-Cov2 3CLPro in complex with Compound 33 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6R)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPW
 
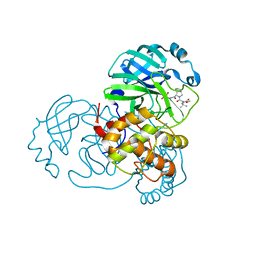 | | Structure of SARS-Cov2 3CLPro in complex with Compound 34 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6S)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
5ZZ3
 
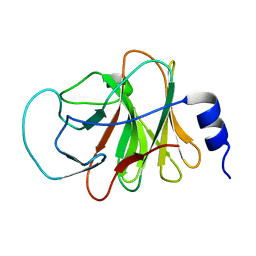 | | Crystal structure of intracellular B30.2 domain of BTN3A3 | | Descriptor: | Butyrophilin, subfamily 3, member A3 isoform b variant | | Authors: | Yang, Y.Y, Li, X, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-05-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
5ZKL
 
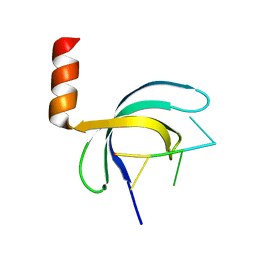 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
5ZKM
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA TCTTCC | | Descriptor: | DNA (5'-D(P*TP*CP*TP*TP*CP*C)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
7DEO
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Spike protein S1, ... | | Authors: | Fu, D, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
