7X4C
 
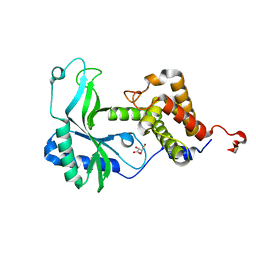 | | Native CD-NTase EfCdnE | | Descriptor: | EfCdnE, L(+)-TARTARIC ACID | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4F
 
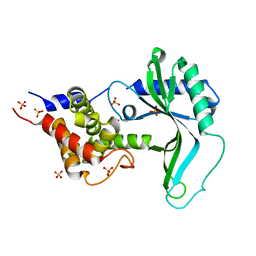 | | Native CD-NTase LpCdnE | | Descriptor: | Cyclic dipyrimidine nucleotide synthase, SULFATE ION | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4T
 
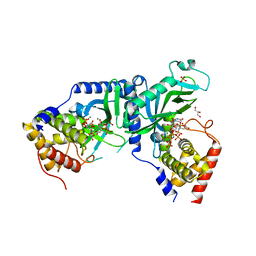 | | LpCdnE UMPNPP Mg complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, Cyclic dipyrimidine nucleotide synthase, GLYCEROL, ... | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4G
 
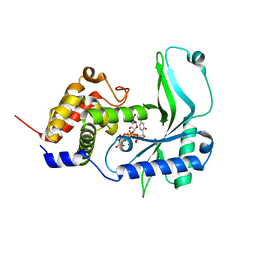 | | CD-NTase ClCdnE in complex with substrate UTP | | Descriptor: | ClCdnE, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7XY7
 
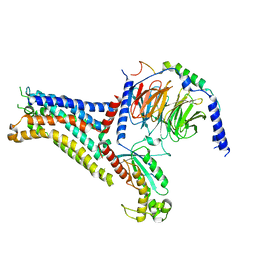 | | Adenosine receptor bound to a non-selective agonist in complex with a G protein obtained by cryo-EM | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | Deposit date: | 2022-05-31 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
7XY6
 
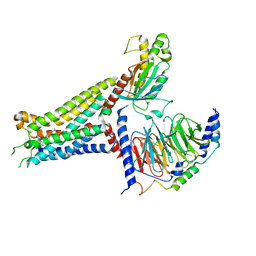 | | Adenosine receptor bound to an agonist in complex with G protein obtained by cryo-EM | | Descriptor: | 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2-yl]sulfanylethanamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | Deposit date: | 2022-05-31 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
7EJU
 
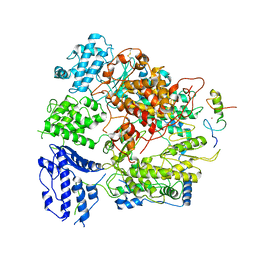 | | Junin virus(JUNV) RNA polymerase L complexed with Z protein | | Descriptor: | MAGNESIUM ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Chen, Y. | | Deposit date: | 2021-04-02 | | Release date: | 2021-07-07 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for recognition and regulation of arenavirus polymerase L by Z protein.
Nat Commun, 12, 2021
|
|
7XVN
 
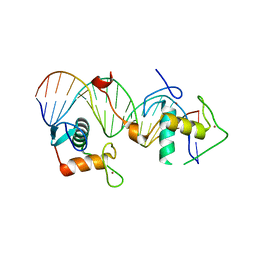 | | Structural basis for DNA recognition feature of retinoid-related orphan receptors | | Descriptor: | DNA (5'-D(P*CP*AP*TP*GP*AP*CP*CP*TP*AP*CP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*GP*GP*TP*CP*AP*GP*TP*AP*GP*GP*TP*CP*AP*TP*G)-3'), Nuclear receptor ROR-gamma, ... | | Authors: | Chen, Y, Jiang, L. | | Deposit date: | 2022-05-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural characterization of the DNA binding mechanism of retinoic acid-related orphan receptor gamma.
Structure, 32, 2024
|
|
7CWE
 
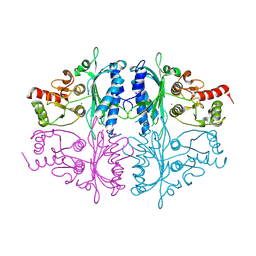 | | Human Fructose-1,6-bisphosphatase 1 in APO R-state | | Descriptor: | Fructose-1,6-bisphosphatase 1, MAGNESIUM ION | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-28 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human Fructose-1,6-bisphosphatase 1 in APO R-state
To Be Published
|
|
7YA2
 
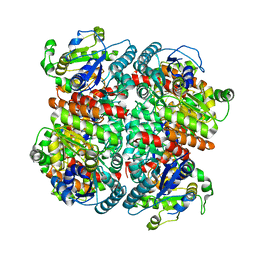 | |
7EQT
 
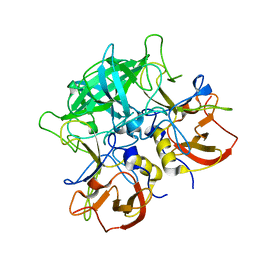 | |
7EQS
 
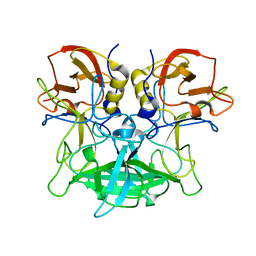 | |
7EQW
 
 | |
7ER1
 
 | |
7ER0
 
 | |
7V4K
 
 | |
7V4J
 
 | |
7V4I
 
 | |
7V4L
 
 | |
7V4H
 
 | |
1FGL
 
 | | Cyclophilin A complexed with a fragment of HIV-1 GAG protein | | Descriptor: | CYCLOPHILIN A, HIV-1 GAG PROTEIN | | Authors: | Zhao, Y, Chen, Y, Schutkowski, M, Fischer, G, Ke, H. | | Deposit date: | 1996-11-18 | | Release date: | 1997-04-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclophilin A complexed with a fragment of HIV-1 gag protein: insights into HIV-1 infectious activity.
Structure, 5, 1997
|
|
4V8U
 
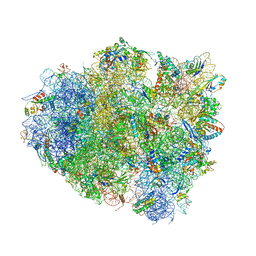 | | Crystal Structure of 70S Ribosome with Both Cognate tRNAs in the E and P Sites Representing an Authentic Elongation Complex. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.G, Feng, S, Chen, Y. | | Deposit date: | 2012-08-28 | | Release date: | 2014-07-09 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex.
PLoS ONE, 8, 2013
|
|
3BVD
 
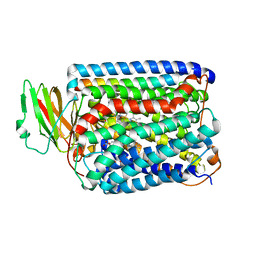 | | Structure of Surface-engineered Cytochrome ba3 Oxidase from Thermus thermophilus under Xenon Pressure, 100psi 5min | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Luna, V.M, Chen, Y, Fee, J.A, Stout, C.D. | | Deposit date: | 2008-01-07 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Crystallographic Studies of Xe and Kr Binding within the Large Internal Cavity of Cytochrome ba3 from Thermus thermophilus: Structural Analysis and Role of Oxygen Transport Channels in the Heme-Cu Oxidases.
Biochemistry, 47, 2008
|
|
3C5V
 
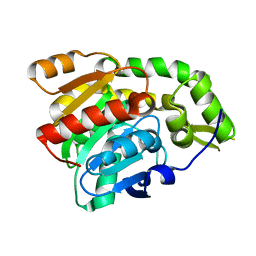 | | PP2A-specific methylesterase apo form (PME) | | Descriptor: | Protein phosphatase methylesterase 1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
2FS1
 
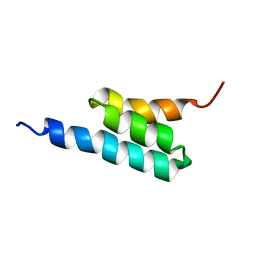 | | solution structure of PSD-1 | | Descriptor: | PSD-1 | | Authors: | He, Y, Rozak, D.A, Sari, N, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2006-01-20 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and stability variation in bacterial albumin binding modules: implications for species specificity.
Biochemistry, 45, 2006
|
|
