8IVZ
 
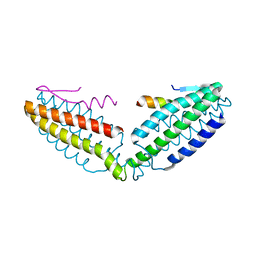 | | Crystal structure of talin R7 in complex with KANK1 KN motif | | Descriptor: | KN motif and ankyrin repeat domains 1, Talin-1 | | Authors: | Xu, Y, Li, K, Wei, Z, Cong, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
5YBU
 
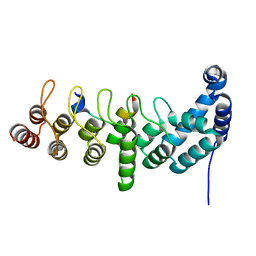 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
8AS9
 
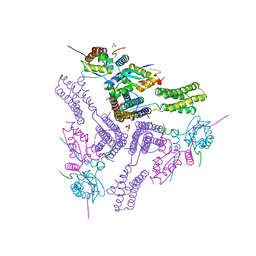 | | Crystal structure of the talin-KANK1 complex | | Descriptor: | B-cell lymphoma 6 protein, GLYCEROL, KN-motif NCoR1 BBD fusion,Nuclear receptor corepressor 1, ... | | Authors: | Zacharchenko, T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis of the talin-KANK1 interaction that coordinates the actin and microtubule cytoskeletons at focal adhesions.
Open Biology, 13, 2023
|
|
5YBJ
 
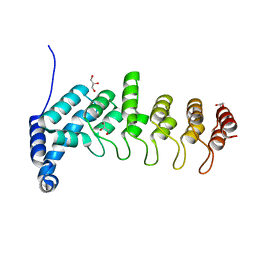 | | Structure of apo KANK1 ankyrin domain | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBV
 
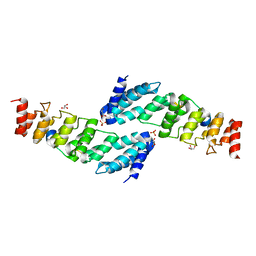 | | The structure of the KANK2 ankyrin domain with the KIF21A peptide | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 2, Kinesin-like protein KIF21A, ... | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
4HBD
 
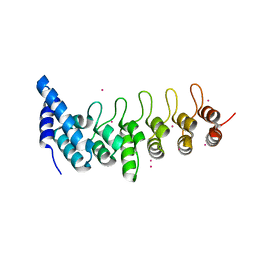 | | Crystal structure of KANK2 ankyrin repeats | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 2, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Tempel, W, Cerovina, T, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-27 | | Release date: | 2012-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of KANK2 ankyrin repeats
To be Published
|
|
5YAY
 
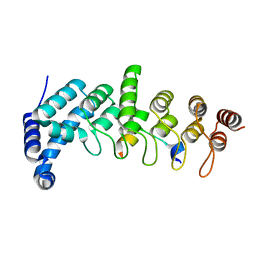 | | Crystal structure of KANK1/KIF21A complex | | Descriptor: | KN motif and ankyrin repeat domains 1, Kinesin-like protein KIF21A | | Authors: | Wei, Z, Pan, W. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.
J. Biol. Chem., 293, 2018
|
|
4E0L
 
 | | FYLLYYT segment from human Beta 2 Microglobulin (62-68) displayed on 54-membered macrocycle scaffold | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide FYLLYYT(ORN)KN(HAO)SA(ORN) | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6NHE
 
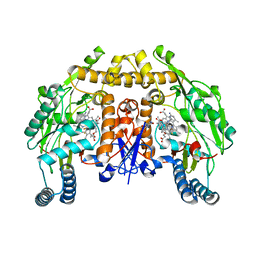 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(5-(2-((2S,4R)-4-ethoxy-1-methylpyrrolidin-2-yl)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(5-{2-[(2S,4R)-4-ethoxy-1-methylpyrrolidin-2-yl]ethyl}-2,3-difluorophenyl)ethyl]-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
6NHB
 
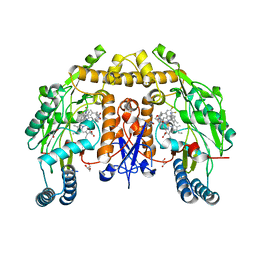 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(5-(2-((2S,4R)-4-ethoxy-1-methylpyrrolidin-2-yl)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(5-{2-[(2S,4R)-4-ethoxy-1-methylpyrrolidin-2-yl]ethyl}-2,3-difluorophenyl)ethyl]-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
7DDX
 
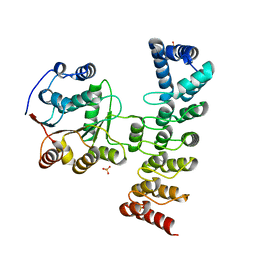 | | Crystal structure of KANK1 S1179F mutant in complex wtih eIF4A1 | | Descriptor: | Eukaryotic initiation factor 4A-I, GLYCEROL, KN motif and ankyrin repeat domains 1, ... | | Authors: | Pan, W, Xu, Y, Wei, Z. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nephrotic-syndrome-associated mutation of KANK2 induces pathologic binding competition with physiological interactor KIF21A.
J.Biol.Chem., 297, 2021
|
|
6TLH
 
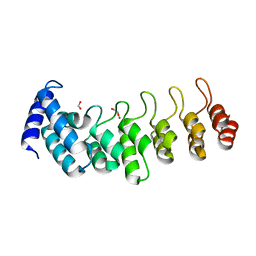 | |
6TMD
 
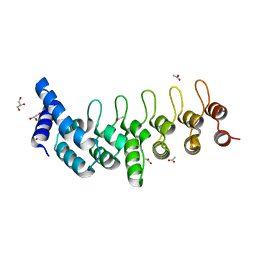 | |
5YAZ
 
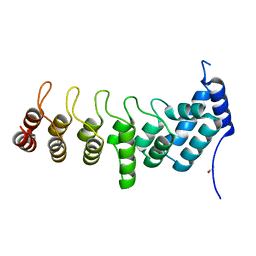 | | Crystal structure of the ANKRD domain of KANK1 | | Descriptor: | ACETATE ION, KN motif and ankyrin repeat domains 1 | | Authors: | Wei, Z, Pan, W. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.
J. Biol. Chem., 293, 2018
|
|
8IW0
 
 | | Crystal structure of the KANK1/liprin-beta1 complex | | Descriptor: | Liprin-beta-1,KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Zhang, J, Chen, S, Wei, Z, Yu, C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
3J3Q
 
 | |
1IW7
 
 | |
1QZV
 
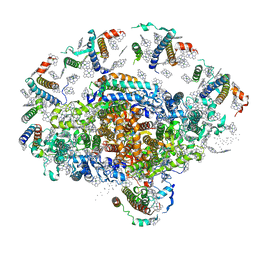 | | Crystal structure of plant photosystem I | | Descriptor: | CHLOROPHYLL A, IRON/SULFUR CLUSTER, PHYLLOQUINONE, ... | | Authors: | Ben-Shem, A, Frolow, F, Nelson, N. | | Deposit date: | 2003-09-18 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.44 Å) | | Cite: | Crystal structure of plant photosystem I.
Nature, 426, 2003
|
|
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7EZX
 
 | |
2A69
 
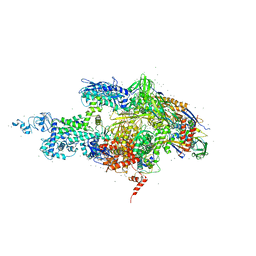 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A68
 
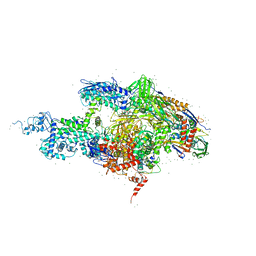 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
3J3Y
 
 | |
8QO1
 
 | | Asymmetric structure of the Borrelia bacteriophage BB1 procapsid, 3D class 3 | | Descriptor: | Cytosolic protein, DUF228 domain-containing protein, Decoration protein P03, ... | | Authors: | Rumnieks, J, Fuzik, T, Tars, K. | | Deposit date: | 2023-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (9.76 Å) | | Cite: | Structure of the Borrelia Bacteriophage phi BB1 Procapsid.
J.Mol.Biol., 435, 2023
|
|
8QO0
 
 | | Asymmetric structure of the Borrelia bacteriophage BB1 procapsid, 3D class 2 | | Descriptor: | Cytosolic protein, DUF228 domain-containing protein, Decoration protein P03, ... | | Authors: | Rumnieks, J, Fuzik, T, Tars, K. | | Deposit date: | 2023-09-27 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (10.62 Å) | | Cite: | Structure of the Borrelia Bacteriophage phi BB1 Procapsid.
J.Mol.Biol., 435, 2023
|
|
