4W93
 
 | | Human pancreatic alpha-amylase in complex with montbretin A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Montbretin A, ... | | Authors: | Williams, L.K, Caner, S, Brayer, G.D. | | Deposit date: | 2014-08-27 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif.
Nat.Chem.Biol., 11, 2015
|
|
8CGT
 
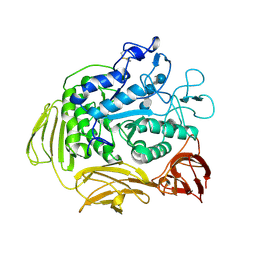 | | STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED WITH A THIO-MALTOHEXAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN-GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose | | Authors: | Schmidt, A.K, Schulz, G.E. | | Deposit date: | 1998-09-27 | | Release date: | 1998-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the gamma-cyclodextrin production.
Eur.J.Biochem., 255, 1998
|
|
8SDB
 
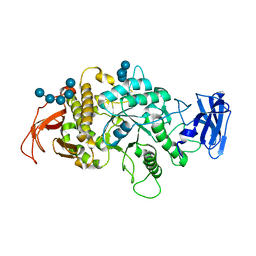 | | Crystal Structure of E.Coli Branching Enzyme in complex with malto-octose | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Bingham, C.R, Nayebi, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Maltooctaose-Bound Escherichia coli Branching Enzyme Suggests a Mechanism for Donor Chain Specificity.
Molecules, 28, 2023
|
|
4X9Y
 
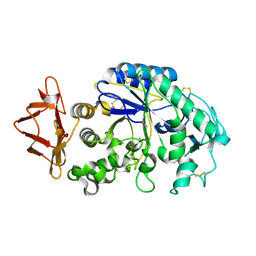 | |
4LPC
 
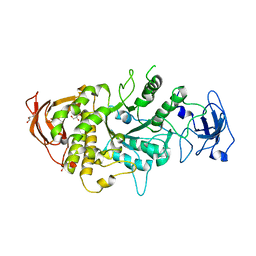 | |
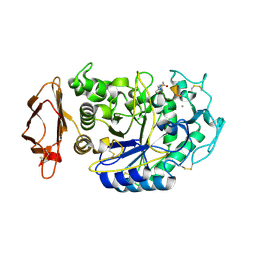
5KEZ
 
 | |
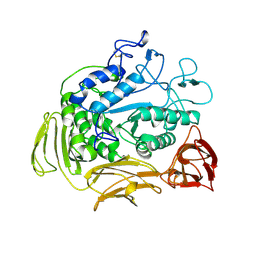
7CGT
 
 | |
6Z8L
 
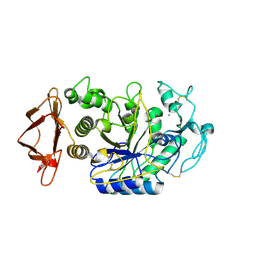 | | Alpha-Amylase in complex with probe fragments | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.40000367 Å) | | Cite: | Enhancing glycan stability via site-selective fluorination: modulating substrate orientation by molecular design.
Chem Sci, 12, 2020
|
|
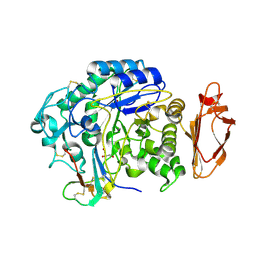
4X0N
 
 | |
4LQ1
 
 | |
9CGT
 
 | | STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED WITH A THIO-MALTOPENTAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN-GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-1-thio-alpha-D-glucopyranose | | Authors: | Schmidt, A.K, Schulz, G.E. | | Deposit date: | 1998-09-27 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the gamma-cyclodextrin production.
Eur.J.Biochem., 255, 1998
|
|
6CGT
 
 | | HOXA COMPLEX OF CYCLODEXTRIN GLYCOSYLTRANSFERASE MUTANT | | Descriptor: | 4-amino-4,6-dideoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Parsiegla, G, Schulz, G.E. | | Deposit date: | 1998-06-06 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the gamma-cyclodextrin production.
Eur.J.Biochem., 255, 1998
|
|
7XSY
 
 | |
7ML5
 
 | | Structure of the Starch Branching Enzyme I (BEI) complexed with maltododecaose from Oryza sativa L | | Descriptor: | Isoform 2 of 1,4-alpha-glucan-branching enzyme, chloroplastic/amyloplastic, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Nayebi Gavgani, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2021-04-27 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structural explanation for the mechanism and specificity of plant branching enzymes I and IIb.
J.Biol.Chem., 298, 2021
|
|
2QMK
 
 | | Human pancreatic alpha-amylase complexed with nitrite | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRITE ION, ... | | Authors: | Williams, L.K, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-07-16 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
1QHP
 
 | | FIVE-DOMAIN ALPHA-AMYLASE FROM BACILLUS STEAROTHERMOPHILUS, MALTOSE COMPLEX | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Dauter, Z, Dauter, M, Brzozowski, A.M, Christensen, S, Borchert, T.V, Beier, L, Wilson, K.S, Davies, G.J. | | Deposit date: | 1999-05-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of Novamyl, the five-domain "maltogenic" alpha-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7A resolution.
Biochemistry, 38, 1999
|
|
1QHO
 
 | | FIVE-DOMAIN ALPHA-AMYLASE FROM BACILLUS STEAROTHERMOPHILUS, MALTOSE/ACARBOSE COMPLEX | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Dauter, Z, Dauter, M, Brzozowski, A.M, Christensen, S, Borchert, T.V, Beier, L, Wilson, K.S, Davies, G.J. | | Deposit date: | 1999-05-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of Novamyl, the five-domain "maltogenic" alpha-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7A resolution.
Biochemistry, 38, 1999
|
|
3L2L
 
 | | X-ray Crystallographic Analysis of Pig Pancreatic Alpha-Amylase with Limit Dextrin and Oligosaccharide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | X-ray crystallographic analyses of pig pancreatic alpha-amylase with limit dextrin, oligosaccharide, and alpha-cyclodextrin.
Biochemistry, 49, 2010
|
|
3L2M
 
 | | X-ray Crystallographic Analysis of Pig Pancreatic Alpha-Amylase with Alpha-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | X-ray crystallographic analyses of pig pancreatic alpha-amylase with limit dextrin, oligosaccharide, and alpha-cyclodextrin.
Biochemistry, 49, 2010
|
|
6WNU
 
 | |
6WNI
 
 | |
8CQG
 
 | | Crystal Structure of a Chimeric Alpha-Amylase from Pseudoalteromonas Haloplanktis | | Descriptor: | 1,2-ETHANEDIOL, Alpha-amylase, CALCIUM ION, ... | | Authors: | Skagseth, S, Lund, B.A, Griese, J.J, van der Ent, F, Aqvist, J. | | Deposit date: | 2023-03-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Computational design of the temperature optimum of an enzyme reaction.
Sci Adv, 9, 2023
|
|
8CQF
 
 | | Crystal Structure of a Chimeric Alpha-Amylase from Pseudoalteromonas Haloplanktis Complexed with Rearranged Acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-1,5-anhydro-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Skagseth, S, Griese, J.J, Lund, B.A, van der Ent, F, Aqvist, J. | | Deposit date: | 2023-03-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of the temperature optimum of an enzyme reaction.
Sci Adv, 9, 2023
|
|
6OCN
 
 | |
8OR6
 
 | |
