6DMX
 
 | | HBZ56 in complex with KIX and c-Myb | | Descriptor: | BZIP factor, CREB-binding protein, Transcriptional activator Myb | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DNQ
 
 | | HBZ77 in complex with KIX and c-Myb | | Descriptor: | 1,2-ETHANEDIOL, BZIP factor, CREB-binding protein, ... | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1HBZ
 
 | | Catalase from Micrococcus lysodeikticu | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Murshudov, G.N, Grebenko, A.I, Barynin, V.V, Braningen, J, Wilson, K.S, Dauter, Z, Melik-Adamyan, W.R. | | Deposit date: | 2001-04-24 | | Release date: | 2001-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-Dimensional Structure of Catalase from Micrococcus Lysodeikticus at 1.5A Resolution
FEBS Lett., 312, 1992
|
|
3HBZ
 
 | |
4HBZ
 
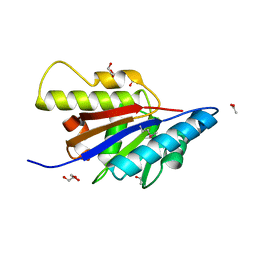 | | The Structure of Putative Phosphohistidine Phosphatase SixA from Nakamurella multipartitia. | | Descriptor: | ACETIC ACID, GLYCEROL, Putative phosphohistidine phosphatase, ... | | Authors: | Cuff, M.E, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of Putative Phosphohistidine Phosphatase SixA from Nakamurella multipartitia.
TO BE PUBLISHED
|
|
5HBZ
 
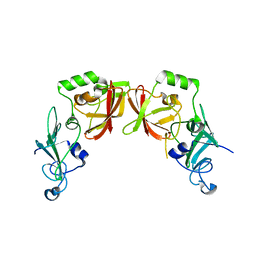 | |
2HBZ
 
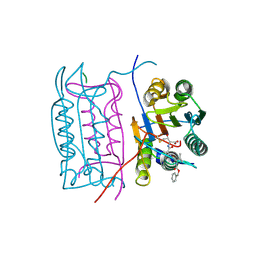 | |
6HBZ
 
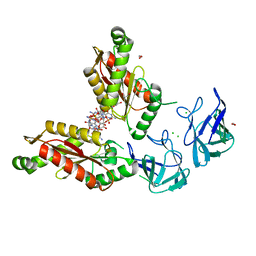 | | Bdellovibrio bacteriovorus DgcB Full-length | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, ... | | Authors: | Lovering, A.L, Meek, R.W. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis for activation of a diguanylate cyclase required for bacterial predation in Bdellovibrio.
Nat Commun, 10, 2019
|
|
5HC1
 
 | |
6HC0
 
 | |
6HC1
 
 | |
2HBR
 
 | |
2HBY
 
 | |
2HBQ
 
 | |
2QN7
 
 | | Glycogen Phosphorylase b in complex with N-4-hydroxybenzoyl-N'-4-beta-D-glucopyranosyl urea | | Descriptor: | Glycogen phosphorylase, muscle form, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Tiraidis, K, Alexacou, K.-M, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | N-(4-substituted-benzoyl)-N'-(beta-D-glucopyranosyl)ureas, inhibitors of glycogen phosphorylase: synthesis, kinetic and crystallographic evaluation
To be Published
|
|
3W4U
 
 | | Human zeta-2 beta-2-s hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit beta, Hemoglobin subunit zeta, ... | | Authors: | Safo, M.K, Ko, T.-P, Russell, J.E. | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of fully liganded Hb zeta 2 beta 2(s) trapped in a tense conformation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1JEB
 
 | | Chimeric Human/Mouse Carbonmonoxy Hemoglobin (Human Zeta2 / Mouse Beta2) | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN BETA-SINGLE CHAIN, HEMOGLOBIN ZETA CHAIN, ... | | Authors: | Kidd, R.D, Russell, J.E, Watmough, N.J, Baker, E.N, Brittain, T. | | Deposit date: | 2001-06-17 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of beta chains in the control of the hemoglobin oxygen binding function: chimeric human/mouse proteins, structure, and function.
Biochemistry, 40, 2001
|
|
5OBP
 
 | | PCE reductive dehalogenase from S. multivorans with 6-hydroxybenzimidazolyl norcobamide cofactor | | Descriptor: | 6-hydroxybenzimidazolyl-norcobamide, BENZAMIDINE, GLYCEROL, ... | | Authors: | Keller, S, Kunze, C, Bommer, M, Paetz, C, Menezes, R.C, Svatos, A, Dobbek, H, Schubert, T. | | Deposit date: | 2017-06-28 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Selective Utilization of Benzimidazolyl-Norcobamides as Cofactors by the Tetrachloroethene Reductive Dehalogenase of Sulfurospirillum multivorans.
J. Bacteriol., 200, 2018
|
|
5OBI
 
 | | PCE reductive dehalogenase from S. multivorans with 5-METHOXYBENZIMIDAZOLYL-NORCOBAMIDE cofactor | | Descriptor: | 5-Methoxybenzimidazolyl-norcobamide, BENZAMIDINE, GLYCEROL, ... | | Authors: | Keller, S, Kunze, C, Bommer, M, Paetz, C, Menezes, R.C, Svatos, A, Dobbek, H, Schubert, T. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Selective Utilization of Benzimidazolyl-Norcobamides as Cofactors by the Tetrachloroethene Reductive Dehalogenase of Sulfurospirillum multivorans.
J. Bacteriol., 200, 2018
|
|
1XWG
 
 | | Human GST A1-1 T68E mutant | | Descriptor: | Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Novotny, M, Grehn, L, Olin, B, Madsen, D, Wahlberg, M, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2004-11-01 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1JAO
 
 | | COMPLEX OF 3-MERCAPTO-2-BENZYLPROPANOYL-ALA-GLY-NH2 WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), N-[(2S)-2-benzyl-3-sulfanylpropanoyl]-L-alanylglycinamide, ... | | Authors: | Grams, F, Reinemer, P, Powers, J.C, Kleine, T, Piper, M, Tschesche, H, Huber, R, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design.
Eur.J.Biochem., 228, 1995
|
|
5M04
 
 | | Structure of ObgE from Escherichia coli | | Descriptor: | GTPase ObgE/CgtA, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Gkekas, S, Singh, R.K, Versees, W. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J. Biol. Chem., 292, 2017
|
|
