2FHT
 
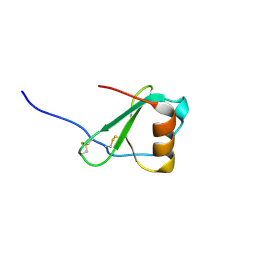 | | Crystal Structure of Viral Macrophage Inflammatory Protein-II | | Descriptor: | Viral macrophage inflammatory protein-II | | Authors: | Li, Y, Liu, D, Cao, R, Kumar, S, Dong, C.Z, wilson, S.R, Gao, Y.G, Huang, Z. | | Deposit date: | 2005-12-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chemically synthesized vMIP-II.
Proteins, 67, 2007
|
|
1A15
 
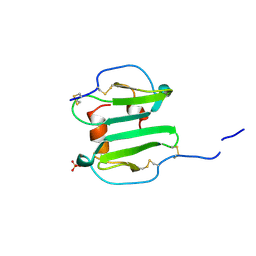 | | SDF-1ALPHA | | Descriptor: | STROMAL DERIVED FACTOR-1ALPHA, SULFATE ION | | Authors: | Dealwis, C.G, Fernandez, E.J, Lolis, E. | | Deposit date: | 1997-12-22 | | Release date: | 1998-08-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1alpha, a potent ligand for the HIV-1 "fusin" coreceptor.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6WZK
 
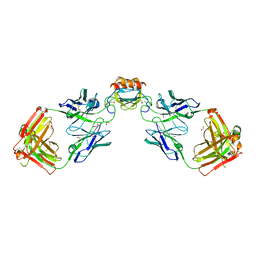 | | LY3041658 Fab bound to CXCL3 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, C-X-C motif chemokine 3, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
1B2T
 
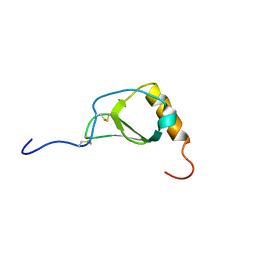 | |
6WZL
 
 | | LY3041658 Fab bound to CXCL7 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, C-X-C motif chemokine 7, LY3041658 Fab heavy chain, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
6WZM
 
 | | LY3041658 Fab bound to CXCL8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-8, LY3041658 Fab heavy chain, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
6WZJ
 
 | | LY3041658 Fab bound to CXCL2 | | Descriptor: | C-X-C motif chemokine 2, LY3041658 Fab Heavy Chain, LY3041658 Fab Light Chain | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
1B53
 
 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
1B50
 
 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, 10 STRUCTURES | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
1BO0
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN-3, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (MONOCYTE CHEMOATTRACTANT PROTEIN-3) | | Authors: | Kwon, D, Lee, D, Sykes, B.D, Kim, K.-S. | | Deposit date: | 1998-08-10 | | Release date: | 1999-10-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of a monomeric chemokine: monocyte chemoattractant protein-3.
FEBS Lett., 395, 1996
|
|
6XMN
 
 | |
1B3A
 
 | | TOTAL CHEMICAL SYNTHESIS AND HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE POTENT ANTI-HIV PROTEIN AOP-RANTES | | Descriptor: | PENTYLOXYAMINO-ACETALDEHYDE, PROTEIN (RANTES), SULFATE ION | | Authors: | Wilken, J, Hoover, D, Thompson, D.A, Barlow, P.N, Mcsparron, H, Picard, L, Wlodawer, A, Lubkowski, J, Kent, S.B.H. | | Deposit date: | 1998-12-07 | | Release date: | 1999-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Total chemical synthesis and high-resolution crystal structure of the potent anti-HIV protein AOP-RANTES.
Chem.Biol., 6, 1999
|
|
2R3Z
 
 | | Crystal structure of mouse IP-10 | | Descriptor: | Small-inducible cytokine B10 | | Authors: | Jabeen, T, Leonard, P, Jamaluddin, H, Acharya, K.R. | | Deposit date: | 2007-08-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of mouse IP-10, a chemokine
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2RA4
 
 | |
2SDF
 
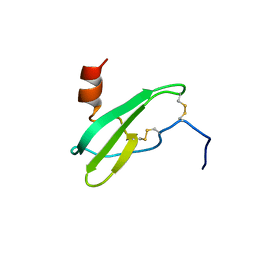 | | SOLUTION NMR STRUCTURE OF STROMAL CELL-DERIVED FACTOR-1 (SDF-1), 30 STRUCTURES | | Descriptor: | STROMAL CELL-DERIVED FACTOR-1 | | Authors: | Crump, M.P, Rajarathnam, K, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1998-03-07 | | Release date: | 1998-06-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1.
EMBO J., 16, 1997
|
|
2VXW
 
 | |
2X6L
 
 | |
2X69
 
 | |
2X6G
 
 | |
5COY
 
 | | Crystal structure of CC chemokine 5 (CCL5) | | Descriptor: | C-C motif chemokine 5, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-07-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5COR
 
 | |
5DNF
 
 | | Crystal structure of CC chemokine 5 (CCL5) oligomer in complex with heparin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, C-C motif chemokine 5, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4R9W
 
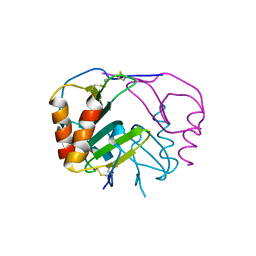 | | Crystal structure of platelet factor 4 complexed with fondaparinux | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Platelet factor 4 | | Authors: | Cai, Z, Zhu, Z, Liu, Q, Greene, M.I. | | Deposit date: | 2014-09-08 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic description of the immune complex involved in heparin-induced thrombocytopenia.
Nat Commun, 6, 2015
|
|
4R9Y
 
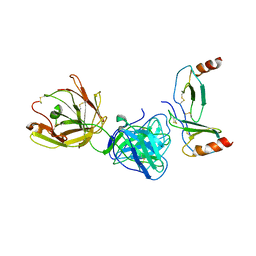 | | Crystal structure of KKOFab in complex with platelet factor 4 | | Descriptor: | Platelet factor 4, Platelet factor 4 antibody KKO heavy chain, Platelet factor 4 antibody KKO light chain | | Authors: | Cai, Z, Zhu, Z, Liu, Q, Greene, M.I. | | Deposit date: | 2014-09-08 | | Release date: | 2015-12-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.11 Å) | | Cite: | Atomic description of the immune complex involved in heparin-induced thrombocytopenia.
Nat Commun, 6, 2015
|
|
4RAU
 
 | |
