8CHH
 
 | |
4LN2
 
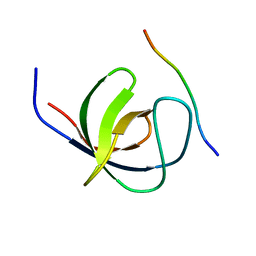 | | The second SH3 domain from CAP/Ponsin in complex with proline rich peptide from Vinculin | | Descriptor: | Sorbin and SH3 domain-containing protein 1, proline rich peptide | | Authors: | Zhao, D, Li, F, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2013-07-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
8CF4
 
 | |
8CHG
 
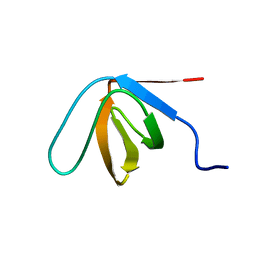 | |
1QWF
 
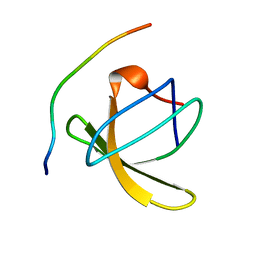 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND VSL12 | | Descriptor: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC, VAL-SER-LEU-ALA-ARG-ARG-PRO-LEU-PRO-PRO-LEU-PRO | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1QWE
 
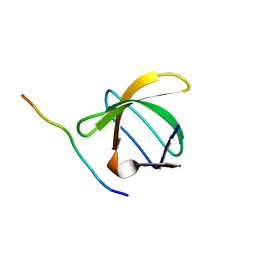 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND APP12 | | Descriptor: | ALA-PRO-PRO-LEU-PRO-PRO-ARG-ASN-ARG-PRO-ARG-LEU, TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
7Y4N
 
 | | Insight into the C-terminal SH3 domain mediated binding of Drosophila Drk to Sos and Dos | | Descriptor: | Growth factor receptor-bound protein 2 | | Authors: | Pooppadi, M.S, Ikeya, T, Sugasawa, H, Watanabe, R, Mishima, M, Inomata, K, Ito, Y. | | Deposit date: | 2022-06-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insight into the C-terminal SH3 domain mediated binding of Drosophila Drk to Sos and Dos.
Biochem.Biophys.Res.Commun., 625, 2022
|
|
7NET
 
 | |
7NES
 
 | |
7NER
 
 | |
7YXW
 
 | | Structure of the p22phox A200G mutant in complex with p47phox peptide | | Descriptor: | Cytochrome b-245 light chain, Neutrophil cytosol factor 1 | | Authors: | Cukier, C.D, Vuillard, L.M, Komjati, B, Szlavik, Z. | | Deposit date: | 2022-02-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting NOX2 via p47/phox-p22/phox Inhibition with Novel Triproline Mimetics
Acs Med.Chem.Lett., 13, 2022
|
|
2YUO
 
 | | Solution structure of the SH3 domain of mouse RUN and TBC1 domain containing 3 | | Descriptor: | RUN and TBC1 domain containing 3 | | Authors: | Abe, H, Tochio, N, Miyamoto, K, Saito, K, Sasagawa, A, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of mouse RUN and TBC1 domain containing 3
To be Published
|
|
6SDF
 
 | | N-terminal SH3 domain of Grb2 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Growth factor receptor-bound protein 2 | | Authors: | Bolgov, A.A, Korban, S.A, Luzik, D.A, Rogacheva, O.N, Zhemkov, V.A, Kim, M, Skrynnikov, N.R, Bezprozvanny, I.B. | | Deposit date: | 2019-07-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the SH3 domain of growth factor receptor-bound protein 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2YUQ
 
 | | Solution structure of the SH3 domain of human Tyrosine-protein kinase ITK/TSK | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Abe, H, Miyamoto, K, Tochio, N, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of human Tyrosine-protein kinase ITK/TSK
To be Published
|
|
8KDX
 
 | |
2YUP
 
 | | Solution structure of the second SH3 domain of human Vinexin | | Descriptor: | Vinexin | | Authors: | Abe, H, Tochio, N, Miyamoto, K, Saito, K, Sasagawa, A, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second SH3 domain of human Vinexin
To be Published
|
|
2YT6
 
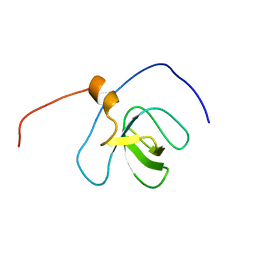 | | Solution structure of the SH3_1 domain of Yamaguchi sarcoma viral (v-yes) oncogene homolog 1 | | Descriptor: | Adult male urinary bladder cDNA, RIKEN full-length enriched library, clone:9530076O17 product:Yamaguchi sarcoma viral (v-yes) oncogene homolog | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3_1 domain of Yamaguchi sarcoma viral (v-yes) oncogene homolog 1
To be Published
|
|
2A36
 
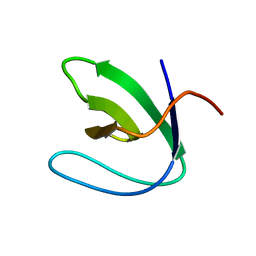 | | Solution structure of the N-terminal SH3 domain of DRK | | Descriptor: | Protein E(sev)2B | | Authors: | Forman-Kay, J.D, Bezsonova, I, Singer, A, Choy, W.-Y, Tollinger, M. | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
2A08
 
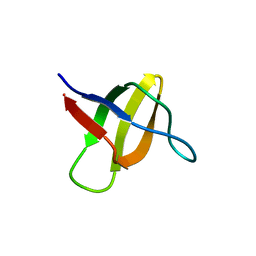 | | Structure of the yeast YHH6 SH3 domain | | Descriptor: | Hypothetical 41.8 kDa protein in SPO13-ARG4 intergenic region | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Lehmann, F, Zou, P, Wilmanns, M. | | Deposit date: | 2005-06-16 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 3-D proteome of yeast SH3 domains
To be Published
|
|
1ZX6
 
 | | High-resolution crystal structure of yeast Pin3 SH3 domain | | Descriptor: | Ypr154wp | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2005-06-07 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of yeast SH3 domains
To be Published
|
|
2A37
 
 | | Solution structure of the T22G mutant of N-terminal SH3 domain of DRK (DRKN SH3 DOMAIN) | | Descriptor: | Protein E(sev)2B | | Authors: | Bezsonova, I, Singer, A, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
7TZK
 
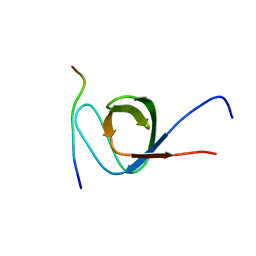 | | EPS8 SH3 Domain with NleH1 PxxDY Motif | | Descriptor: | Epidermal growth factor receptor kinase substrate 8, T3SS secreted effector NleH homolog | | Authors: | Grishin, A.M, Cygler, M. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Targeting of microvillus protein Eps8 by the NleH effector kinases from enteropathogenic E. coli
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7CFZ
 
 | | SH3 domain of NADPH oxidase activator 1 | | Descriptor: | NADPH oxidase activator 1 | | Authors: | Kim, M, Park, J.H, Attri, P, Lee, W. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural modification of NADPH oxidase activator (Noxa 1) by oxidative stress: An experimental and computational study.
Int.J.Biol.Macromol., 163, 2020
|
|
3M0P
 
 | |
3M0S
 
 | |
