-Search query
-Search result
Showing 1 - 50 of 163 items for (author: lara & s)

EMDB-42528: 
CryoEM structure of A/Perth/16/2009 H3 in complex with flu HA central stem VH1-18 antibody UCA6

EMDB-42529: 
CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody 09-1B12

EMDB-42530: 
CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody UCA6_N55T

EMDB-42531: 
CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-15 days post immunization

EMDB-42532: 
CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization

EMDB-42533: 
CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-15 days post-immunization

EMDB-42534: 
CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-28 days post immunization

EMDB-42535: 
CryoEM structure of A/Perth/16/2009 H3

EMDB-42536: 
CryoEM map of A/Shanghai/1/2013 H7 HA

PDB-8ut3: 
CryoEM structure of A/Perth/16/2009 H3 in complex with flu HA central stem VH1-18 antibody UCA6

PDB-8ut4: 
CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody 09-1B12

PDB-8ut5: 
CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody UCA6_N55T

PDB-8ut6: 
CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-15 days post immunization

PDB-8ut7: 
CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization

PDB-8ut8: 
CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-15 days post-immunization

PDB-8ut9: 
CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-28 days post immunization

EMDB-17350: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution

PDB-8p1i: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution

EMDB-27641: 
The structure of the interleukin 11 signalling complex, truncated gp130

EMDB-27642: 
The structure of the IL-11 signalling complex, with full-length extracellular gp130

PDB-8dps: 
The structure of the interleukin 11 signalling complex, truncated gp130

PDB-8dpt: 
The structure of the IL-11 signalling complex, with full-length extracellular gp130
EMDB-18149: 
GBP1 bound by 14-3-3sigma

PDB-8q4l: 
GBP1 bound by 14-3-3sigma

EMDB-17576: 
SARS-CoV-2 S-protein:D614G mutant in 1-up conformation

EMDB-17578: 
SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor

PDB-8p99: 
SARS-CoV-2 S-protein:D614G mutant in 1-up conformation

PDB-8p9y: 
SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor

EMDB-41624: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+ condition

EMDB-41628: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+-free condition

EMDB-41629: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+ condition (C1 map)

EMDB-41630: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+-free condition (C1 map)

PDB-8tul: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+ condition

PDB-8tup: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+-free condition

EMDB-15403: 
Structure of the secreted Yersinia entomophaga Tc toxin YenTc

EMDB-15404: 
Tomogram of Yersinia entomophaga in the pre-secretory state

EMDB-15405: 
Tomogram of Yersinia entomophaga in the post-holin state
EMDB-15406: 
Tomogram of Yersinia entomophaga in the post-endolysin state
EMDB-15407: 
Tomogram of Yersinia entomophaga in the post-spanin state

EMDB-41046: 
Subtomogram average structure of the injectisome
EMDB-41047: 
Intermembrane Tether between SCV and Mitochondria

EMDB-15700: 
Type 3 secretion system needle complex of Shigella flexneri

EMDB-15701: 
Outer membrane secretin pore of the type 3 secretion system of Shigella flexneri

EMDB-15702: 
Inner membrane ring and secretin N0 N1 domains of the type 3 secretion system of Shigella flexneri

EMDB-15703: 
Inner membrane ring of the type 3 secretion system of Shigella flexneri

PDB-8axk: 
Type 3 secretion system export apparatus core, inner rod and needle of Shigella flexneri

PDB-8axl: 
Outer membrane secretin pore of the type 3 secretion system of Shigella flexneri

PDB-8axn: 
Inner membrane ring and secretin N0 N1 domains of the type 3 secretion system of Shigella flexneri
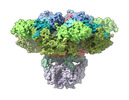
EMDB-27982: 
Structure of the full-length IP3R1 channel determined at high Ca2+
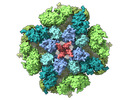
EMDB-27983: 
Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
