[English] 日本語
 Yorodumi
Yorodumi- EMDB-42530: CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody UCA6_N55T | |||||||||
 Map data Map data | Mich15H1 UCA6_N55T_sharp | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Flu Hemagglutinin / Central stem epitope / VH1-18 /  VIRAL PROTEIN / VIRAL PROTEIN /  VIRAL PROTEIN-IMMUNE SYSTEM complex VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology information viral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane viral envelope / virion attachment to host cell / host cell plasma membrane / virion membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /    Influenza A virus Influenza A virus | |||||||||
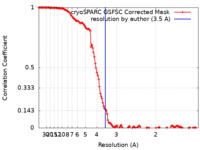
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Huang J / Han J / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2024 Journal: Immunity / Year: 2024Title: Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses. Authors: Rashmi Ray / Faez Amokrane Nait Mohamed / Daniel P Maurer / Jiachen Huang / Berk A Alpay / Larance Ronsard / Zhenfei Xie / Julianna Han / Monica Fernandez-Quintero / Quynh Anh Phan / Rebecca ...Authors: Rashmi Ray / Faez Amokrane Nait Mohamed / Daniel P Maurer / Jiachen Huang / Berk A Alpay / Larance Ronsard / Zhenfei Xie / Julianna Han / Monica Fernandez-Quintero / Quynh Anh Phan / Rebecca L Ursin / Mya Vu / Kathrin H Kirsch / Thavaleak Prum / Victoria C Rosado / Thalia Bracamonte-Moreno / Vintus Okonkwo / Julia Bals / Caitlin McCarthy / Usha Nair / Masaru Kanekiyo / Andrew B Ward / Aaron G Schmidt / Facundo D Batista / Daniel Lingwood /   Abstract: Broadly neutralizing antibodies (bnAbs) targeting the hemagglutinin (HA) stem of influenza A viruses (IAVs) tend to be effective against either group 1 or group 2 viral diversity. In rarer cases, ...Broadly neutralizing antibodies (bnAbs) targeting the hemagglutinin (HA) stem of influenza A viruses (IAVs) tend to be effective against either group 1 or group 2 viral diversity. In rarer cases, intergroup protective bnAbs can be generated by human antibody paratopes that accommodate the conserved glycan differences between the group 1 and group 2 stems. We applied germline-engaging nanoparticle immunogens to elicit a class of cross-group bnAbs from physiological precursor frequency within a humanized mouse model. Cross-group protection depended on the presence of the human bnAb precursors within the B cell repertoire, and the vaccine-expanded antibodies enriched for an N55T substitution in the CDRH2 loop, a hallmark of the bnAb class. Structurally, this single mutation introduced a flexible fulcrum to accommodate glycosylation differences and could alone enable cross-group protection. Thus, broad IAV immunity can be expanded from the germline repertoire via minimal antigenic input and an exceptionally simple antibody development pathway. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42530.map.gz emd_42530.map.gz | 203.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42530-v30.xml emd-42530-v30.xml emd-42530.xml emd-42530.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42530_fsc.xml emd_42530_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_42530.png emd_42530.png | 52.8 KB | ||
| Filedesc metadata |  emd-42530.cif.gz emd-42530.cif.gz | 6.3 KB | ||
| Others |  emd_42530_half_map_1.map.gz emd_42530_half_map_1.map.gz emd_42530_half_map_2.map.gz emd_42530_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42530 http://ftp.pdbj.org/pub/emdb/structures/EMD-42530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42530 | HTTPS FTP |
-Related structure data
| Related structure data |  8ut5MC  8ut3C  8ut4C  8ut6C  8ut7C  8ut8C  8ut9C  8uwaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42530.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42530.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mich15H1 UCA6_N55T_sharp | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.725 Å | ||||||||||||||||||||||||||||||||||||
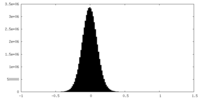
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Mich15H1 UCA6 N55T halfA
| File | emd_42530_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mich15H1 UCA6_N55T_halfA | ||||||||||||
| Projections & Slices |
| ||||||||||||
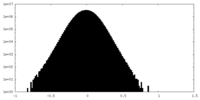
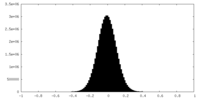
| Density Histograms |
-Half map: Mich15H1 UCA6 N55T halfA
| File | emd_42530_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mich15H1 UCA6_N55T_halfA | ||||||||||||
| Projections & Slices |
| ||||||||||||
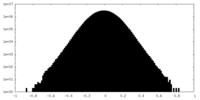
| Density Histograms |
- Sample components
Sample components
-Entire : An UCA6 Fab complex with one protomer of a H3 trimer
| Entire | Name: An UCA6 Fab complex with one protomer of a H3 trimer |
|---|---|
| Components |
|
-Supramolecule #1: An UCA6 Fab complex with one protomer of a H3 trimer
| Supramolecule | Name: An UCA6 Fab complex with one protomer of a H3 trimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Hemagglutinin HA1 chain
| Macromolecule | Name: Hemagglutinin HA1 chain / type: protein_or_peptide / ID: 1 / Details: A/Michigan/45/2015 H1 HA1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 36.018465 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ADTLCIGYHA NNSTDTVDTV LEKNVTVTHS VNLLEDKHNG KLCKLRGVAP LHLGKCNIAG WILGNPECES LSTASSWSYI VETSNSDNG TCYPGDFINY EELREQLSSV SSFERFEIFP KTSSWPNHDS NKGVTAACPH AGAKSFYKNL IWLVKKGNSY P KLNQSYIN ...String: ADTLCIGYHA NNSTDTVDTV LEKNVTVTHS VNLLEDKHNG KLCKLRGVAP LHLGKCNIAG WILGNPECES LSTASSWSYI VETSNSDNG TCYPGDFINY EELREQLSSV SSFERFEIFP KTSSWPNHDS NKGVTAACPH AGAKSFYKNL IWLVKKGNSY P KLNQSYIN DKGKEVLVLW GIHHPSTTAD QQSLYQNADA YVFVGTSRYS KKFKPEIATR PKVRDQEGRM NYYWTLVEPG DK ITFEATG NLVVPRYAFT MERNAGSGII ISDTPVHDCN TTCQTPEGAI NTSLPFQNIH PITIGKCPKY VKSTKLRLAT GLR NVPS UniProtKB:  Hemagglutinin Hemagglutinin |
-Macromolecule #2: Hemagglutinin HA2 chain
| Macromolecule | Name: Hemagglutinin HA2 chain / type: protein_or_peptide / ID: 2 / Details: A/Michigan/45/2015 H1 HA2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 28.359492 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: IQSRGLFGAI AGFIEGGWTG MVDGWYGYHH QNEQGSGYAA DLKSTQNAID KITNKVNSVI EKMNTQFTAV GKEFNHLEKR IENLNKKVD DGFLDIWTYN AELLVLLENE RTLDYHDSNV KNLYEKVRNQ LKNNAKEIGN GCFEFYHKCD NTCMESVKNG T YDYPKYSE ...String: IQSRGLFGAI AGFIEGGWTG MVDGWYGYHH QNEQGSGYAA DLKSTQNAID KITNKVNSVI EKMNTQFTAV GKEFNHLEKR IENLNKKVD DGFLDIWTYN AELLVLLENE RTLDYHDSNV KNLYEKVRNQ LKNNAKEIGN GCFEFYHKCD NTCMESVKNG T YDYPKYSE EAKLNREKID GVKGALEVLF QGPGSHHHHH HHHLGGSGYI PEAPRDGQAY VRKDGEWVLL STFLGSGGGL ND IFEAQKI EWH UniProtKB:  Hemagglutinin Hemagglutinin |
-Macromolecule #3: UCA6_N55T HC Fv
| Macromolecule | Name: UCA6_N55T HC Fv / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.893534 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKASGYTFT SYGISWVRQA PGQGLEWMGW ISAYTGNTNY AQKLQGRVTM TTDTSTSTAY MELRSLRSD DTAVYYCARG LLQGVVILDS YYYTMDVWGQ GTTVTVSS |
-Macromolecule #4: UCA6_N55T LC Fv
| Macromolecule | Name: UCA6_N55T LC Fv / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.858149 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQSVS SSYLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQYGSSPRWT FGQGTKLEIK |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 17 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)