-Search query
-Search result
Showing 1 - 50 of 463 items for (author: brown & ng)

EMDB-43889: 
Chlamydomonas reinhardtii mastigoneme (constituent map 1)

EMDB-43890: 
Chlamydomonas reinhardtii mastigoneme (constituent map 2)

EMDB-43891: 
Chlamydomonas reinhardtii mastigoneme (constituent map 3)

EMDB-43892: 
Composite cryo-EM map of the Chlamydomonas reinhardtii mastigoneme

PDB-9b4h: 
Chlamydomonas reinhardtii mastigoneme filament

EMDB-41363: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5

PDB-8tl6: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5

EMDB-19067: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1).

EMDB-19076: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon, mRNA and P-site tRNA (structure 2).

EMDB-19077: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon and EF-Tu(GDP) (structure 3).

PDB-8rd8: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1).

PDB-8rdv: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon, mRNA and P-site tRNA (structure 2).

PDB-8rdw: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon and EF-Tu(GDP) (structure 3).
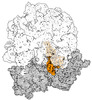
EMDB-43074: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4)
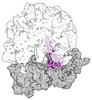
EMDB-43075: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Rv2629 (Balon) (Structure 5)
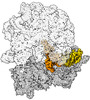
EMDB-43076: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Composite structure 6)

EMDB-43077: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Structure 6)

EMDB-43078: 
Hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) bound to Mycobacterium smegmatis 70S ribosome, from focused 3D classification and refinement (Structure 6)

PDB-8v9j: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4)

PDB-8v9k: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Rv2629 (Balon) (Structure 5)

PDB-8v9l: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Composite structure 6)

EMDB-29723: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate, 30S focused map

EMDB-29724: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate consensus, 30S focused

EMDB-29833: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate, 30S focused

EMDB-29834: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate, 50S focused from 30S subtracted

EMDB-29842: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate, 50S focused and 30S subtracted

EMDB-29681: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms

EMDB-29687: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate

EMDB-29688: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate

EMDB-29689: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate

EMDB-29844: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate, 50S focused and 30S subtracted

PDB-8g2u: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms

PDB-8g31: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate

PDB-8g34: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate

PDB-8g38: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate

EMDB-17187: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm

PDB-8otz: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm
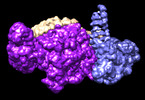
EMDB-41097: 
cryoEM structure of Smc5/6 5mer

EMDB-41098: 
Smc5/6 8mer

PDB-8t8e: 
cryoEM structure of Smc5/6 5mer

PDB-8t8f: 
Smc5/6 8mer

EMDB-40334: 
Human OCT1 (Apo) in inward-open conformation

EMDB-40335: 
Human OCT1 bound to diltiazem in inward-open conformation

EMDB-40336: 
Human OCT1 bound to fenoterol in inward-open conformation

EMDB-40337: 
Human OCT1 bound to metformin in inward-open conformation

EMDB-40339: 
Human OCT1 bound to thiamine in inward-open conformation

PDB-8sc1: 
Human OCT1 (Apo) in inward-open conformation

PDB-8sc2: 
Human OCT1 bound to diltiazem in inward-open conformation

PDB-8sc3: 
Human OCT1 bound to fenoterol in inward-open conformation

PDB-8sc4: 
Human OCT1 bound to metformin in inward-open conformation
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
