[English] 日本語
 Yorodumi
Yorodumi- PDB-8v9j: Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8v9j | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4) | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords |  RIBOSOME / RIBOSOME /  cryo-EM / cryo-EM /  mycobacteria / mycobacteria /  hibernation / Msmeg1130 / Balon hibernation / Msmeg1130 / Balon | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationlarge ribosomal subunit rRNA binding / large ribosomal subunit /  regulation of translation / small ribosomal subunit / regulation of translation / small ribosomal subunit /  5S rRNA binding / 5S rRNA binding /  transferase activity / transferase activity /  tRNA binding / tRNA binding /  rRNA binding / rRNA binding /  ribosome / structural constituent of ribosome ...large ribosomal subunit rRNA binding / large ribosomal subunit / ribosome / structural constituent of ribosome ...large ribosomal subunit rRNA binding / large ribosomal subunit /  regulation of translation / small ribosomal subunit / regulation of translation / small ribosomal subunit /  5S rRNA binding / 5S rRNA binding /  transferase activity / transferase activity /  tRNA binding / tRNA binding /  rRNA binding / rRNA binding /  ribosome / structural constituent of ribosome / ribosome / structural constituent of ribosome /  ribonucleoprotein complex / ribonucleoprotein complex /  translation / translation /  mRNA binding / zinc ion binding / mRNA binding / zinc ion binding /  metal ion binding / metal ion binding /  cytoplasm cytoplasmSimilarity search - Function | ||||||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | ||||||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.1 Å cryo EM / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | Rybak, M.Y. / Helena-Bueno, K. / Hill, C.H. / Melnikov, S.V. / Gagnon, M.G. | ||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: A new family of bacterial ribosome hibernation factors. Authors: Karla Helena-Bueno / Mariia Yu Rybak / Chinenye L Ekemezie / Rudi Sullivan / Charlotte R Brown / Charlotte Dingwall / Arnaud Baslé / Claudia Schneider / James P R Connolly / James N Blaza / ...Authors: Karla Helena-Bueno / Mariia Yu Rybak / Chinenye L Ekemezie / Rudi Sullivan / Charlotte R Brown / Charlotte Dingwall / Arnaud Baslé / Claudia Schneider / James P R Connolly / James N Blaza / Bálint Csörgő / Patrick J Moynihan / Matthieu G Gagnon / Chris H Hill / Sergey V Melnikov /    Abstract: To conserve energy during starvation and stress, many organisms use hibernation factor proteins to inhibit protein synthesis and protect their ribosomes from damage. In bacteria, two families of ...To conserve energy during starvation and stress, many organisms use hibernation factor proteins to inhibit protein synthesis and protect their ribosomes from damage. In bacteria, two families of hibernation factors have been described, but the low conservation of these proteins and the huge diversity of species, habitats and environmental stressors have confounded their discovery. Here, by combining cryogenic electron microscopy, genetics and biochemistry, we identify Balon, a new hibernation factor in the cold-adapted bacterium Psychrobacter urativorans. We show that Balon is a distant homologue of the archaeo-eukaryotic translation factor aeRF1 and is found in 20% of representative bacteria. During cold shock or stationary phase, Balon occupies the ribosomal A site in both vacant and actively translating ribosomes in complex with EF-Tu, highlighting an unexpected role for EF-Tu in the cellular stress response. Unlike typical A-site substrates, Balon binds to ribosomes in an mRNA-independent manner, initiating a new mode of ribosome hibernation that can commence while ribosomes are still engaged in protein synthesis. Our work suggests that Balon-EF-Tu-regulated ribosome hibernation is a ubiquitous bacterial stress-response mechanism, and we demonstrate that putative Balon homologues in Mycobacteria bind to ribosomes in a similar fashion. This finding calls for a revision of the current model of ribosome hibernation inferred from common model organisms and holds numerous implications for how we understand and study ribosome hibernation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8v9j.cif.gz 8v9j.cif.gz | 3.3 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8v9j.ent.gz pdb8v9j.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  8v9j.json.gz 8v9j.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v9/8v9j https://data.pdbj.org/pub/pdb/validation_reports/v9/8v9j ftp://data.pdbj.org/pub/pdb/validation_reports/v9/8v9j ftp://data.pdbj.org/pub/pdb/validation_reports/v9/8v9j | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 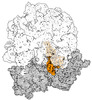 43074MC  8rd8C  8rdvC  8rdwC  8v9kC  8v9lC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-RNA chain , 5 types, 5 molecules avyAB
| #1: RNA chain |  Mass: 495373.656 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: GenBank: 118168627 |
|---|---|
| #22: RNA chain | Mass: 873.540 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 |
| #23: RNA chain | Mass: 24531.672 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 |
| #25: RNA chain |  Mass: 1026283.438 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: GenBank: 118168627 |
| #26: RNA chain |  Mass: 38061.816 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: GenBank: 118168627 |
-30S Ribosomal Protein ... , 20 types, 20 molecules bcdefghijklmnopqrstu
| #2: Protein |  / Small ribosomal subunit protein uS2 / Small ribosomal subunit protein uS2Mass: 30145.230 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QVB8 |
|---|---|
| #3: Protein |  Mass: 30191.227 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSD7 |
| #4: Protein |  Mass: 23415.787 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSL7 |
| #5: Protein |  / Small ribosomal subunit protein uS5 / Small ribosomal subunit protein uS5Mass: 21946.090 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSG6 |
| #6: Protein |  Mass: 10991.637 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0A2U9Q0X2 |
| #7: Protein |  Mass: 17660.375 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QS97 |
| #8: Protein |  / Small ribosomal subunit protein uS8 / Small ribosomal subunit protein uS8Mass: 14492.638 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSG3 |
| #9: Protein |  Mass: 16794.365 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSP9 |
| #10: Protein |  Mass: 11454.313 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSD0 |
| #11: Protein |  Mass: 14671.762 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSL6 |
| #12: Protein |  Mass: 13896.366 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QS96 |
| #13: Protein |  Mass: 14249.619 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSL5 |
| #14: Protein |  Mass: 6976.409 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSG2 |
| #15: Protein |  Mass: 10368.097 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QVQ3 |
| #16: Protein |  Mass: 16795.207 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QV37 |
| #17: Protein |  Mass: 11127.002 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSE0 |
| #18: Protein |  Mass: 9524.188 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0R7F7 |
| #19: Protein |  Mass: 10800.602 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0QSD5 |
| #20: Protein |  Mass: 9556.104 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / References: UniProt: A0R102 |
| #21: Protein/peptide |  Mass: 4164.300 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 |
+50S Ribosomal Protein ... , 33 types, 33 molecules CDEFGHIJKLMNOPQRSTUVWXYZ123456...
-Protein / Non-polymers , 2 types, 3 molecules z

| #24: Protein | Mass: 41390.672 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: MSMEG_1130 Source: (gene. exp.)  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria)Strain: MC2155 / Gene: MSMEG_1130 / Plasmid: pET28-SMT3-MSMEG_1130 / Production host:   Escherichia coli BL21(DE3) (bacteria) / References: UniProt: A0QRI6 Escherichia coli BL21(DE3) (bacteria) / References: UniProt: A0QRI6 |
|---|---|
| #60: Chemical |
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4) Type: RIBOSOME / Entity ID: #1-#59 / Source: NATURAL |
|---|---|
| Molecular weight | Value: 2.6 MDa / Experimental value: NO |
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Buffer solution | pH: 7.5 Details: 20 mM HEPES-KOH, 60 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
| Specimen support | Grid material: GOLD / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R2/1 |
Vitrification | Instrument: LEICA EM GP / Cryogen name: ETHANE / Humidity: 85 % / Chamber temperature: 295 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 105000 X / Nominal defocus max: 2000 nm / Nominal defocus min: 800 nm / Cs Bright-field microscopy / Nominal magnification: 105000 X / Nominal defocus max: 2000 nm / Nominal defocus min: 800 nm / Cs : 2.7 mm / C2 aperture diameter: 100 µm / Alignment procedure: COMA FREE : 2.7 mm / C2 aperture diameter: 100 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 40.1 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 11031 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry : C1 (asymmetric) : C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 302401 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 5o61 Accession code: 5o61 / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller







 PDBj
PDBj































