+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
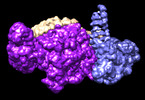
| Title | Smc5/6 8mer | |||||||||
 Map data Map data | sharp map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Smc / Nse /  Condensin / Condensin /  Cohesin / Cohesin /  DNA binding / DNA binding /  DNA damage / DNA damage /  DNA repair / DNA repair /  DNA BINDING PROTEIN DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationSmc5-Smc6 complex / resolution of DNA recombination intermediates / DNA double-strand break attachment to nuclear envelope / chromosome separation / SUMOylation of DNA damage response and repair proteins / chromatin looping /  recombinational repair / recombinational repair /  regulation of telomere maintenance / protein sumoylation / double-strand break repair via homologous recombination ...Smc5-Smc6 complex / resolution of DNA recombination intermediates / DNA double-strand break attachment to nuclear envelope / chromosome separation / SUMOylation of DNA damage response and repair proteins / chromatin looping / regulation of telomere maintenance / protein sumoylation / double-strand break repair via homologous recombination ...Smc5-Smc6 complex / resolution of DNA recombination intermediates / DNA double-strand break attachment to nuclear envelope / chromosome separation / SUMOylation of DNA damage response and repair proteins / chromatin looping /  recombinational repair / recombinational repair /  regulation of telomere maintenance / protein sumoylation / double-strand break repair via homologous recombination / regulation of telomere maintenance / protein sumoylation / double-strand break repair via homologous recombination /  single-stranded DNA binding / site of double-strand break / damaged DNA binding / single-stranded DNA binding / site of double-strand break / damaged DNA binding /  chromosome, telomeric region / chromosome, telomeric region /  DNA repair / DNA repair /  ATP hydrolysis activity / ATP hydrolysis activity /  mitochondrion / mitochondrion /  ATP binding / ATP binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Saccharomyces cerevisiae W303 (yeast) Saccharomyces cerevisiae W303 (yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.8 Å cryo EM / Resolution: 4.8 Å | |||||||||
 Authors Authors | Yu Y / Patel DJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions. Authors: Shibai Li / You Yu / Jian Zheng / Victoria Miller-Browne / Zheng Ser / Huihui Kuang / Dinshaw J Patel / Xiaolan Zhao /   Abstract: The Smc5/6 complex (Smc5/6) is important for genome replication and repair in eukaryotes. Its cellular functions are closely linked to the ATPase activity of the Smc5 and Smc6 subunits. This activity ...The Smc5/6 complex (Smc5/6) is important for genome replication and repair in eukaryotes. Its cellular functions are closely linked to the ATPase activity of the Smc5 and Smc6 subunits. This activity requires the dimerization of the motor domains of the two SMC subunits and is regulated by the six non-SMC subunits (Nse1 to Nse6). Among the NSEs, Nse5 and Nse6 form a stable subcomplex (Nse5-6) that dampens the ATPase activity of the complex. However, the underlying mechanisms and biological significance of this regulation remain unclear. Here, we address these issues using structural and functional studies. We determined cryo-EM structures of the yeast Smc5/6 derived from complexes consisting of either all eight subunits or a subset of five subunits. Both structures reveal that Nse5-6 associates with Smc6's motor domain and the adjacent coiled-coil segment, termed the neck region. Our structural analyses reveal that this binding is compatible with motor domain dimerization but results in dislodging the Nse4 subunit from the Smc6 neck. As the Nse4-Smc6 neck interaction favors motor domain engagement and thus ATPase activity, Nse6's competition with Nse4 can explain how Nse5-6 disfavors ATPase activity. Such regulation could in principle differentially affect Smc5/6-mediated processes depending on their needs of the complex's ATPase activity. Indeed, mutagenesis data in cells provide evidence that the Nse6-Smc6 neck interaction is important for the resolution of DNA repair intermediates but not for replication termination. Our results thus provide a molecular basis for how Nse5-6 modulates the ATPase activity and cellular functions of Smc5/6. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41098.map.gz emd_41098.map.gz | 203.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41098-v30.xml emd-41098-v30.xml emd-41098.xml emd-41098.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41098.png emd_41098.png | 73.8 KB | ||
| Filedesc metadata |  emd-41098.cif.gz emd-41098.cif.gz | 8 KB | ||
| Others |  emd_41098_half_map_1.map.gz emd_41098_half_map_1.map.gz emd_41098_half_map_2.map.gz emd_41098_half_map_2.map.gz | 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41098 http://ftp.pdbj.org/pub/emdb/structures/EMD-41098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41098 | HTTPS FTP |
-Related structure data
| Related structure data |  8t8fMC  8t8eC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41098.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41098.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharp map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.069 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map B
| File | emd_41098_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_41098_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Smc5/6 8mer
| Entire | Name: Smc5/6 8mer |
|---|---|
| Components |
|
-Supramolecule #1: Smc5/6 8mer
| Supramolecule | Name: Smc5/6 8mer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae W303 (yeast) Saccharomyces cerevisiae W303 (yeast) |
| Molecular weight | Theoretical: 400 kDa/nm |
-Macromolecule #1: DNA repair protein KRE29
| Macromolecule | Name: DNA repair protein KRE29 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae W303 (yeast) Saccharomyces cerevisiae W303 (yeast) |
| Molecular weight | Theoretical: 53.836758 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae W303 (yeast) Saccharomyces cerevisiae W303 (yeast) |
| Sequence | String: MGSVNSSPNE EFETVPDSQI SGFDSPLIPT SVGSYFRDDD DDEKVHPNFI SDPENDSLNS DEEFSSLENS DLNLSGAKAE SGDDFDPIL KRTIISKRKA PSNNEDEEIV KTPRKLVNYV PLKIFNLGDS FDDTITTTVA KLQDLKKEIL DSPRSNKSIV I TSNTVAKS ...String: MGSVNSSPNE EFETVPDSQI SGFDSPLIPT SVGSYFRDDD DDEKVHPNFI SDPENDSLNS DEEFSSLENS DLNLSGAKAE SGDDFDPIL KRTIISKRKA PSNNEDEEIV KTPRKLVNYV PLKIFNLGDS FDDTITTTVA KLQDLKKEIL DSPRSNKSIV I TSNTVAKS ELQKSIKFSG SIPEIYLDVV TKETISDKYK DWHFISKNCH YEQLMDLEMK DTAYSFLFGS SRSQGKVPEF VH LKCPSIT NLLVLFGVNQ EKCNSLKINY EKKENSRYDN LCTIFPVNKM LKFLMYFYSD DDNDDVREFF LKAFICLILD RKV FNAMES DHRLCFKVLE LFNEAHFINS YFEIVDKNDF FLHYRLLQIF PHLQSALLRR RFSEKQGRTE TIQQNIIKEF NEFF DCKNY KNLLYFILTM YGSKFIPFGP KCQVTEYFKD CILDISNETT NDVEISILKG ILNLFSKIR UniProtKB:  DNA repair protein KRE29 DNA repair protein KRE29 |
-Macromolecule #2: Non-structural maintenance of chromosome element 5
| Macromolecule | Name: Non-structural maintenance of chromosome element 5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae W303 (yeast) Saccharomyces cerevisiae W303 (yeast) |
| Molecular weight | Theoretical: 64.079609 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae W303 (yeast) Saccharomyces cerevisiae W303 (yeast) |
| Sequence | String: MDGALINSVL YVSPRNGAHY FVELTEKHLL AFEMLNSMCL LENYDHVLLF LECQFGKSHN LAVIPFDIIL VLFTLSTLSE YYKEPILRA NDPYNTSRET LSRRALKLLQ KYLAILKEFD SEQYNLYDLE LLRCQFFLAI DTLTPKKQKW GFDRFRRTKS E SGVTYRQN ...String: MDGALINSVL YVSPRNGAHY FVELTEKHLL AFEMLNSMCL LENYDHVLLF LECQFGKSHN LAVIPFDIIL VLFTLSTLSE YYKEPILRA NDPYNTSRET LSRRALKLLQ KYLAILKEFD SEQYNLYDLE LLRCQFFLAI DTLTPKKQKW GFDRFRRTKS E SGVTYRQN ASVDPELDQA KTFKNPYRSY ISCLEQRNTI LGNRLLNLKL NEPGEFINMI LWTLSNSLQE STPLFLSSHE IW MPLLEIL IDLFSCRQDY FIQHEVAQNV SKSLFVQRLS ESPLAVFFES LNTRNFANRF SEYVFLNCDY KLPSDNYATP VHP VYNGEN TIVDTYIPTI KCSPLYKSQK SLALRRKLIG SCFKLLLRVP DGHRLITPRI VADDVIQGIS RTLASFNDIL QFKK FFMTE NLSQESYFIP LLAEGTLSEI LKDTQECVVI LTLVENLSDG VSFCNEVIGL VKSKCFAFTE QCSQASYEEA VLNIE KCDV CLLVLLRYLL HLIGTEAILD AKEQLEMLHA IEKNDSGRRQ WAKALNLGND PPLLYPIVSQ MFGVHDKSVI IE UniProtKB: Non-structural maintenance of chromosome element 5 |
-Macromolecule #3: Structural maintenance of chromosomes protein 5
| Macromolecule | Name: Structural maintenance of chromosomes protein 5 / type: protein_or_peptide / ID: 3 Details: Smc5 E1015Q mutation, visible part is from 32-246aa and 883-1093aa. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae W303 (yeast) Saccharomyces cerevisiae W303 (yeast) |
| Molecular weight | Theoretical: 126.23618 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MTSLIDLGRY VERTHHGEDT EPRSKRVKIA KPDLSSFQPG SIIKIRLQDF VTYTLTEFNL SPSLNMIIGP NGSGKSTFVC AVCLGLAGK PEYIGRSKKV EDFIKNGQDV SKIEITLKNS PNVTDIEYID ARDETIKITR IITRSKRRSD YLINDYQVSE S VVKTLVAQ ...String: MTSLIDLGRY VERTHHGEDT EPRSKRVKIA KPDLSSFQPG SIIKIRLQDF VTYTLTEFNL SPSLNMIIGP NGSGKSTFVC AVCLGLAGK PEYIGRSKKV EDFIKNGQDV SKIEITLKNS PNVTDIEYID ARDETIKITR IITRSKRRSD YLINDYQVSE S VVKTLVAQ LNIQLDNLCQ FLSQERVEEF ARLKSVKLLV ETIRSIDASL LDVLDELREL QGNEQSLQKD LDFKKAKIVH LR QESDKLR KSVESLRDFQ NKKGEIELHS QLLPYVKVKD HKEKLNIYKE EYERAKANLR AILKDKKPFA NTKKTLENQV EEL TEKCSL KTDEFLKAKE KINEIFEKLN TIRDEVIKKK NQNEYYRGRT KKLQATIIST KEDFLRSQEI LAQTHLPEKS VFED IDIKR KEIINKEGEI RDLISEIDAK ANAINHEMRS IQRQAESKTK SLTTTDKIGI LNQDQDLKEV RDAVLMVREH PEMKD KILE PPIMTVSAIN AQFAAYLAQC VDYNTSKALT VVDSDSYKLF ANPILDKFKV NLRELSSADT TPPVPAETVR DLGFEG YLS DFITGDKRVM KMLCQTSKIH TIPVSRRELT PAQIKKLITP RPNGKILFKR IIHGNRLVDI KQSAYGSKQV FPTDVSI KQ TNFYQGSIMS NEQKIRIENE IINLKNEYND RKSTLDALSN QKSGYRHELS ELASKNDDIN REAHQLNEIR KKYTMRKS T IETLREKLDQ LKREARKDVS QKIKDIDDQI QQLLLKQRHL LSKMASSMKS LKNCQKELIS TQILQFEAQN MDVSMNDVI GFFNEREADL KSQYEDKKKF VKEMRDTPEF QSWMREIRSY DQDTKEKLNK VAEKYEEEGN FNLSFVQDVL DKLESEIAMV NHDESAVTI LDQVTAELRE LEHTVPQQSK DLETIKAKLK EDHAVLEPKL DDIVSKISAR FARLFNNVGS AGAVRLEKPK D YAEWKIEI MVKFRDNAPL KKLDSHTQSG GERAVSTVLY MIALQEFTSA PFRVVDQINQ GMDSRNERIV HKAMVENACA EN TSQYFLI TPKLLTGLHY HEKMRIHCVM AGSWIPNPSE DPKMIHFGET SNYSFD UniProtKB: Structural maintenance of chromosomes protein 5 |
-Macromolecule #4: Structural maintenance of chromosomes protein 6
| Macromolecule | Name: Structural maintenance of chromosomes protein 6 / type: protein_or_peptide / ID: 4 Details: Smc6 E1048Q mutation with C terminal 2X strep tag, visible part is from 79-286aa and 923-1084aa Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae W303 (yeast) Saccharomyces cerevisiae W303 (yeast) |
| Molecular weight | Theoretical: 128.198742 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MISTTISGKR PIEQVDDELL SLTAQQENEE QQQQRKRRRH QFAPMTQFNS NTLDEDSGFR SSSDVATADQ DNFLEESPSG YIKKVILRN FMCHEHFELE LGSRLNFIVG NNGSGKSAIL TAITIGLGAK ASETNRGSSL KDLIREGCYS AKIILHLDNS K YGAYQQGI ...String: MISTTISGKR PIEQVDDELL SLTAQQENEE QQQQRKRRRH QFAPMTQFNS NTLDEDSGFR SSSDVATADQ DNFLEESPSG YIKKVILRN FMCHEHFELE LGSRLNFIVG NNGSGKSAIL TAITIGLGAK ASETNRGSSL KDLIREGCYS AKIILHLDNS K YGAYQQGI FGNEIIVERI IKRDGPASFS LRSENGKEIS NKKKDIQTVV DYFSVPVSNP MCFLSQDAAR SFLTASTSQD KY SHFMKGT LLQEITENLL YASAIHDSAQ ENMALHLENL KSLKAEYEDA KKLLRELNQT SDLNERKMLL QAKSLWIDVA HNT DACKNL ENEISGIQQK VDEVTEKIRN RQEKIERYTS DGTTIEAQID AKVIYVNEKD SEHQNARELL RDVKSRFEKE KSNQ AEAQS NIDQGRKKVD ALNKTIAHLE EELTKEMGGD KDQMRQELEQ LEKANEKLRE VNNSLVVSLQ DVKNEERDIQ HERES ELRT ISRSIQNKKV ELQNIAKGND TFLMNFDRNM DRLLRTIEQR KNEFETPAIG PLGSLVTIRK GFEKWTRSIQ RAISSS LNA FVVSNPKDNR LFRDIMRSCG IRSNIPIVTY CLSQFDYSKG RAHGNYPTIV DALEFSKPEI ECLFVDLSRI ERIVLIE DK NEARNFLQRN PVNVNMALSL RDRRSGFQLS GGYRLDTVTY QDKIRLKVNS SSDNGTQYLK DLIEQETKEL QNIRDRYE E KLSEVRSRLK EIDGRLKSTK NEMRKTNFRM TELKMNVGKV VDTGILNSKI NERKNQEQAI ASYEAAKEEL GLKIEQIAQ EAQPIKEQYD STKLALVEAQ DELQQLKEDI NSRQSKIQKY KDDTIYYEDK KKVYLENIKK IEVNVAALKE GIQRQIQNAC AFCSKERIE NVDLPDTQEE IKRELDKVSR MIQKAEKSLG LSQEEVIALF EKCRNKYKEG QKKYMEIDEA LNRLHNSLKA R DQNYKNAE KGTCFDADMD FRASLKVRKF SGNLSFIKDT KSLEIYILTT NDEKARNVDT LSGGEKSFSQ MALLLATWKP MR SRIIALD QFDVFMDQVN RKIGTTLIVK KLKDIARTQT IIITPQDIGK IADIDSSGVS IHRMRDPERQ NNSNFYN UniProtKB: Structural maintenance of chromosomes protein 6 |
-Macromolecule #5: Non-structural maintenance of chromosome element 4
| Macromolecule | Name: Non-structural maintenance of chromosome element 4 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae W303 (yeast) Saccharomyces cerevisiae W303 (yeast) |
| Molecular weight | Theoretical: 46.195945 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MSSTVISRKR RNSTVTEPDS SGETRKQKKS RSDEKSSSSK DGDPQLEFKV LQGYRDLESE MHKGRAQVTR TGDIGVAMDN LNAVDSLFN KVIGIKNNGL FAHDARAMVS ISELAQISVR NLKFDDSRSM VNLENIVNSL KRYMLKEHFK LNNIAENRND L TLAADEQS ...String: MSSTVISRKR RNSTVTEPDS SGETRKQKKS RSDEKSSSSK DGDPQLEFKV LQGYRDLESE MHKGRAQVTR TGDIGVAMDN LNAVDSLFN KVIGIKNNGL FAHDARAMVS ISELAQISVR NLKFDDSRSM VNLENIVNSL KRYMLKEHFK LNNIAENRND L TLAADEQS AADQQEESDG DIDRTPDDNH TDKATSSFKA TSMRHSYLQQ FSHYNEFSQF NWFRIGALYN TISKNAPITD HL MGPLSIE KKPRVLTQRR RNNDQVGEKI TAEKITQHSL NSTQQETTPE QVKKCFKKLS KKLGPEGSIN LFKFIIDPNS FSR SIENLF YTSFLIKEGK LLMEHDEEGL PTIKIKQSIS HTDSRSKEIE RQRRRAAHQN HIIFQMDMPT WRKLIKKYNI TSPF LD UniProtKB: Non-structural maintenance of chromosome element 4 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 25 mM Hepes, pH 7.5, 250 mM NaCl, 1 mM DTT |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 53.55 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 145290 |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X