-Search query
-Search result
Showing all 43 items for (author: meir & a)

EMDB-19008: 
Cryo-EM structure of the human mPSF with PAPOA C-terminus peptide (PAPOAc)

PDB-8r8r: 
Cryo-EM structure of the human mPSF with PAPOA C-terminus peptide (PAPOAc)

EMDB-36138: 
CrtSPARTA Octamer bound with guide-target

PDB-8jay: 
CrtSPARTA Octamer bound with guide-target

EMDB-36059: 
Short ago complexed with TIR-APAZ

PDB-8j84: 
Short ago complexed with TIR-APAZ

EMDB-36070: 
SPARTA monomer bound with guide-target, state 2

EMDB-36095: 
CrtSPARTA hetero-dimer bound with guide-target, state 1

EMDB-36114: 
SPARTA dimer bound with guide-target

PDB-8j8h: 
SPARTA monomer bound with guide-target, state 2

PDB-8j9g: 
CrtSPARTA hetero-dimer bound with guide-target, state 1

PDB-8j9p: 
SPARTA dimer bound with guide-target
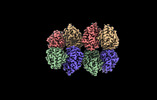
EMDB-28013: 
Cryo-EM structure of the Glutaminase C core filament (fGAC)

PDB-8ec6: 
Cryo-EM structure of the Glutaminase C core filament (fGAC)

EMDB-16144: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the macrocyclic peptide S1B3inL1

PDB-8bon: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the macrocyclic peptide S1B3inL1
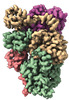
EMDB-28961: 
Cryo-EM structure of the human BCDX2 complex
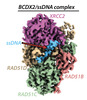
EMDB-29917: 
Cryo-EM structure of a human BCDX2/ssDNA complex

PDB-8faz: 
Cryo-EM structure of the human BCDX2 complex

PDB-8gbj: 
Cryo-EM structure of a human BCDX2/ssDNA complex

EMDB-15860: 
Cryo-EM structure of ribosome-Sec61-TRAP (TRanslocon Associated Protein) translocon complex

PDB-8b5l: 
Cryo-EM structure of ribosome-Sec61-TRAP (TRanslocon Associated Protein) translocon complex

EMDB-15863: 
Cryo-EM structure of ribosome-Sec61 in complex with cyclotriazadisulfonamide derivative CK147

PDB-8b6c: 
Cryo-EM structure of ribosome-Sec61 in complex with cyclotriazadisulfonamide derivative CK147

EMDB-14582: 
Human heparan sulfate polymerase complex EXT1-EXT2

PDB-7zay: 
Human heparan sulfate polymerase complex EXT1-EXT2

EMDB-14867: 
Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded)

EMDB-14870: 
Monomeric PSI of Chlamydomonas reinhardtii at 2.31 A resolution

EMDB-14871: 
Dimeric PSI of Chlamydomonas reinhardtii at 2.97 A resolution

EMDB-14872: 
Plastocyanin bound to PSI of Chlamydomonas reinhardtii

PDB-7zq9: 
Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded)

PDB-7zqc: 
Monomeric PSI of Chlamydomonas reinhardtii at 2.31 A resolution

PDB-7zqd: 
Dimeric PSI of Chlamydomonas reinhardtii at 2.97 A resolution

PDB-7zqe: 
Plastocyanin bound to PSI of Chlamydomonas reinhardtii

EMDB-14185: 
Human RBBP6-bound CPSF complex

EMDB-25164: 
Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP

PDB-7sjr: 
Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP

EMDB-13083: 
L. pneumophila Type IV Coupling Complex (T4CC) with density for DotY N-terminal and middle domains

EMDB-13858: 
L. pneumophila Type IV Coupling Complex (T4CC) computed from WT particles where DotY/Z is missing (referred to as T4CCWTminusYZ at 6.3A)

EMDB-13859: 
L. pneumophila Type IV Coupling Complex (T4CC) computed from a sample where dotY/Z genes were deleted (referred to as T4CCdeltaDotYZ at 15A)

PDB-7ovb: 
L. pneumophila Type IV Coupling Complex (T4CC) with density for DotY N-terminal and middle domains

EMDB-10350: 
Type IV Coupling Complex (T4CC) from L. pneumophila.

PDB-6sz9: 
Type IV Coupling Complex (T4CC) from L. pneumophila.
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
