-Search query
-Search result
Showing 1 - 50 of 65 items for (author: sandeep & n)

EMDB-40650: 
Phosphoinositide phosphate 3 kinase gamma
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40651: 
Phosphoinositide phosphate 3 kinase gamma bound with ATP
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40652: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40653: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40654: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 1
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40655: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 2
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

PDB-8so9: 
Phosphoinositide phosphate 3 kinase gamma
Method: single particle / : Chen CL, Tesmer JJG

PDB-8soa: 
Phosphoinositide phosphate 3 kinase gamma bound with ATP
Method: single particle / : Chen CL, Tesmer JJG

PDB-8sob: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP
Method: single particle / : Chen CL, Tesmer JJG

PDB-8soc: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma
Method: single particle / : Chen CL, Tesmer JJG

PDB-8sod: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 1
Method: single particle / : Chen CL, Tesmer JJG

PDB-8soe: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 2
Method: single particle / : Chen CL, Tesmer JJG

EMDB-29493: 
Cryo-EM structure of human Needle/NAIP/NLRC4 (R288A)
Method: single particle / : Matico RE, Yu X, Miller R, Somani S, Ricketts MD, Kumar N, Steele RA, Medley Q, Berger S, Faustin B, Sharma S

EMDB-29496: 
Cryo-EM structure of full-length human NLRC4 inflammasome with C11 symmetry
Method: single particle / : Matico RE, Yu X, Miller R, Somani S, Ricketts MD, Kumar N, Steele RA, Medley Q, Berger S, Faustin B, Sharma S

EMDB-29498: 
Cryo-EM structure of full-length human NLRC4 inflammasome with C12 symmetry
Method: single particle / : Matico RE, Yu X, Miller R, Somani S, Ricketts MD, Kumar N, Steele RA, Medley Q, Berger S, Faustin B, Sharma S

PDB-8fvu: 
Cryo-EM structure of human Needle/NAIP/NLRC4 (R288A)
Method: single particle / : Matico RE, Yu X, Miller R, Somani S, Ricketts MD, Kumar N, Steele RA, Medley Q, Berger S, Faustin B, Sharma S

PDB-8fw2: 
Cryo-EM structure of full-length human NLRC4 inflammasome with C11 symmetry
Method: single particle / : Matico RE, Yu X, Miller R, Somani S, Ricketts MD, Kumar N, Steele RA, Medley Q, Berger S, Faustin B, Sharma S

PDB-8fw9: 
Cryo-EM structure of full-length human NLRC4 inflammasome with C12 symmetry
Method: single particle / : Matico RE, Yu X, Miller R, Somani S, Ricketts MD, Kumar N, Steele RA, Medley Q, Berger S, Faustin B, Sharma S

EMDB-28910: 
Glycan-Base ConC Env Trimer
Method: single particle / : Olia AS, Kwong PD
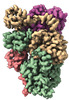
EMDB-28961: 
Cryo-EM structure of the human BCDX2 complex
Method: single particle / : Jia L, Wasmuth EV, Ruben EA, Sung P, Rawal Y, Greene EC, Meir A, Olsen SK
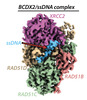
EMDB-29917: 
Cryo-EM structure of a human BCDX2/ssDNA complex
Method: single particle / : Jia L, Wasmuth EV, Ruben EA, Sung P, Rawal Y, Greene EC, Meir A, Olsen SK

PDB-8faz: 
Cryo-EM structure of the human BCDX2 complex
Method: single particle / : Jia L, Wasmuth EV, Ruben EA, Sung P, Rawal Y, Greene EC, Meir A, Olsen SK

PDB-8gbj: 
Cryo-EM structure of a human BCDX2/ssDNA complex
Method: single particle / : Jia L, Wasmuth EV, Ruben EA, Sung P, Rawal Y, Greene EC, Meir A, Olsen SK

EMDB-16232: 
Structure of the TRAP complex with the Sec translocon and a translating ribosome
Method: single particle / : Jaskolowski M, Jomaa A, Gamerdinger M, Shrestha S, Leibundgut M, Deuerling E, Ban N

PDB-8btk: 
Structure of the TRAP complex with the Sec translocon and a translating ribosome
Method: single particle / : Jaskolowski M, Jomaa A, Gamerdinger M, Shrestha S, Leibundgut M, Deuerling E, Ban N
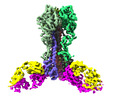
EMDB-27139: 
Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8d21: 
Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-25100: 
Unmethylated Mtb Ribosome 50S with SEQ-9
Method: single particle / : Xing Z, Cui Z

PDB-7sfr: 
Unmethylated Mtb Ribosome 50S with SEQ-9
Method: single particle / : Xing Z, Cui Z, Zhang J, TB Structural Genomics Consortium (TBSGC)

EMDB-25153: 
Human Trio residues 1284-1959 in complex with Rac1
Method: single particle / : Chen CL, Ravala SK, Bandekar SJ, Cash J, Tesmer JJG

PDB-7sj4: 
Human Trio residues 1284-1959 in complex with Rac1
Method: single particle / : Chen CL, Ravala SK, Bandekar SJ, Cash J, Tesmer JJG

EMDB-25905: 
Cryo-EM structure of W6 possum enterovirus
Method: single particle / : Wang I, Jayawardena N

PDB-7thx: 
Cryo-EM structure of W6 possum enterovirus
Method: single particle / : Wang I, Jayawardena N, Strauss M, Bostina M

EMDB-22865: 
CryoEM structure of A2296-methylated Mycobacterium tuberculosis ribosome bound with SEQ-9
Method: single particle / : Cui Z, Zhang J

PDB-7kgb: 
CryoEM structure of A2296-methylated Mycobacterium tuberculosis ribosome bound with SEQ-9
Method: single particle / : Cui Z, Zhang J

EMDB-25792: 
Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

PDB-7tb4: 
Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

EMDB-24663: 
Cryo-EM density map of the outer dynein arm core from bovine tracheal cilia
Method: single particle / : Gui M, Anderson JR, Botsch JJ, Meleppattu S, Singh SK, Zhang Q, Brown A

EMDB-24664: 
Composite cryo-EM density map of the 48-nm repeat doublet microtubule from bovine tracheal cilia
Method: single particle / : Gui M, Anderson JR, Botsch JJ, Meleppattu S, Singh SK, Zhang Q, Brown A

PDB-7rro: 
Structure of the 48-nm repeat doublet microtubule from bovine tracheal cilia
Method: single particle / : Gui M, Anderson JR, Botsch JJ, Meleppattu S, Singh SK, Zhang Q, Brown A

EMDB-23914: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

EMDB-23915: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

PDB-7mlz: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

PDB-7mm0: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

EMDB-23498: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody A23-58.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky Y

EMDB-23499: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody A23-58.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky T

PDB-7lrs: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody A23-58.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky Y

PDB-7lrt: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody A23-58.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky T

EMDB-12018: 
VAR2CSA full ectodomain in present of plCS, DBL1-DBL4
Method: single particle / : Wang KT, Dagil R, Gourdon PE, Salanti A
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
