[English] 日本語
 Yorodumi
Yorodumi- EMDB-27139: Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited,... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 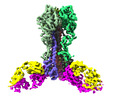 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information viral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane /  membrane membraneSimilarity search - Function | |||||||||
| Biological species |    Influenza A virus / Influenza A virus /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.96 Å cryo EM / Resolution: 3.96 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2023 Journal: Sci Transl Med / Year: 2023Title: An influenza H1 hemagglutinin stem-only immunogen elicits a broadly cross-reactive B cell response in humans. Authors: Sarah F Andrews / Lauren Y Cominsky / Geoffrey D Shimberg / Rebecca A Gillespie / Jason Gorman / Julie E Raab / Joshua Brand / Adrian Creanga / Suprabhath R Gajjala / Sandeep Narpala / ...Authors: Sarah F Andrews / Lauren Y Cominsky / Geoffrey D Shimberg / Rebecca A Gillespie / Jason Gorman / Julie E Raab / Joshua Brand / Adrian Creanga / Suprabhath R Gajjala / Sandeep Narpala / Crystal S F Cheung / Darcy R Harris / Tongqing Zhou / Ingelise Gordon / LaSonji Holman / Floreliz Mendoza / Katherine V Houser / Grace L Chen / John R Mascola / Barney S Graham / Peter D Kwong / Alicia Widge / Lesia K Dropulic / Julie E Ledgerwood / Masaru Kanekiyo / Adrian B McDermott /  Abstract: Current yearly seasonal influenza vaccines primarily induce an antibody response directed against the immunodominant but continually diversifying hemagglutinin (HA) head region. These antibody ...Current yearly seasonal influenza vaccines primarily induce an antibody response directed against the immunodominant but continually diversifying hemagglutinin (HA) head region. These antibody responses provide protection against the vaccinating strain but little cross-protection against other influenza strains or subtypes. To focus the immune response on subdominant but more conserved epitopes on the HA stem that might protect against a broad range of influenza strains, we developed a stabilized H1 stem immunogen lacking the immunodominant head displayed on a ferritin nanoparticle (H1ssF). Here, we evaluated the B cell response to H1ssF in healthy adults ages 18 to 70 in a phase 1 clinical trial (NCT03814720). We observed both a strong plasmablast response and sustained elicitation of cross-reactive HA stem-specific memory B cells after vaccination with H1ssF in individuals of all ages. The B cell response was focused on two conserved epitopes on the H1 stem, with a highly restricted immunoglobulin repertoire unique to each epitope. On average, two-thirds of the B cell and serological antibody response recognized a central epitope on the H1 stem and exhibited broad neutralization across group 1 influenza virus subtypes. The remaining third recognized an epitope near the viral membrane anchor and was largely limited to H1 strains. Together, we demonstrate that an H1 HA immunogen lacking the immunodominant HA head produces a robust and broadly neutralizing HA stem-directed B cell response. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27139.map.gz emd_27139.map.gz | 96.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27139-v30.xml emd-27139-v30.xml emd-27139.xml emd-27139.xml | 26.6 KB 26.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27139.png emd_27139.png | 98.5 KB | ||
| Masks |  emd_27139_msk_1.map emd_27139_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_27139_additional_1.map.gz emd_27139_additional_1.map.gz emd_27139_additional_2.map.gz emd_27139_additional_2.map.gz emd_27139_half_map_1.map.gz emd_27139_half_map_1.map.gz emd_27139_half_map_2.map.gz emd_27139_half_map_2.map.gz | 32.1 MB 13.1 MB 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27139 http://ftp.pdbj.org/pub/emdb/structures/EMD-27139 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27139 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27139 | HTTPS FTP |
-Related structure data
| Related structure data |  8d21MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27139.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27139.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27139_msk_1.map emd_27139_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened map
| File | emd_27139_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: density modified with resolve
| File | emd_27139_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density modified with resolve | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_27139_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_27139_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited,...
| Entire | Name: Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited,...
| Supramolecule | Name: Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: 1B06 Fab with NC99 HA trimer
| Supramolecule | Name: 1B06 Fab with NC99 HA trimer / type: complex / ID: 2 / Chimera: Yes / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
-Macromolecule #1: Hemagglutinin HA2 chain
| Macromolecule | Name: Hemagglutinin HA2 chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 25.263104 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GLFGAIAGFI EGGWTGMVDG WYGYHHQNEQ GSGYAADQKS TQNAINCITN KVNSVIEKMN TQFTAVGKEF NKLERRMENL NKKVDDGFL DIWTYNAELL VLLENERTLD FHDSNVKNLY EKVKSQLKNN AKEIGNGCFE FYHKCNNECM ESVKNGTYDY P KYSEESKL ...String: GLFGAIAGFI EGGWTGMVDG WYGYHHQNEQ GSGYAADQKS TQNAINCITN KVNSVIEKMN TQFTAVGKEF NKLERRMENL NKKVDDGFL DIWTYNAELL VLLENERTLD FHDSNVKNLY EKVKSQLKNN AKEIGNGCFE FYHKCNNECM ESVKNGTYDY P KYSEESKL NREKIDGVSG RLVPRGSPGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGHH HHHH |
-Macromolecule #2: Hemagglutinin HA1 chain
| Macromolecule | Name: Hemagglutinin HA1 chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 36.407852 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DTICIGYHAN NSTDTVDTVC EKNVTVTHSV NLLEDSHNGK LCLLKGIAPL QLGNCSVAGW ILGNPECELL ISKESWSYIV ETPNPENGT CYPGYFADYE ELREQLSSVS SFERFEIFPK ESSWPNHTVT GVSASCSHNG KSSFYRNLLW LTGKNGLYPN L SKSYVNNK ...String: DTICIGYHAN NSTDTVDTVC EKNVTVTHSV NLLEDSHNGK LCLLKGIAPL QLGNCSVAGW ILGNPECELL ISKESWSYIV ETPNPENGT CYPGYFADYE ELREQLSSVS SFERFEIFPK ESSWPNHTVT GVSASCSHNG KSSFYRNLLW LTGKNGLYPN L SKSYVNNK EKEVLVLWGV HHPPNIGNQR ALYHTENAYV SVVSSHYSRR FTPEIAKRPK VRDQEGRINY YWTLLEPGDT II FEANGNL IAPWYAFALS RGFGSGIITS NAPMDECDAK CQTPQGAINS SLPFQNVHPV TIGECPKYVR SAKLRMVTGL RNI PSIQSR |
-Macromolecule #3: 1B06 Heavy Chain
| Macromolecule | Name: 1B06 Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.499133 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLLESGGG LVRPGGSLRL SCKASGFSFS RHGLSWVRQA PGKGLEWVSS ISGSSLSRYY ADSVKGRFTI SRDNSQNTLY LQMNSLRVE DTAVFYCVKD RLDTAVPRGW FDSWGQGILV TVSS |
-Macromolecule #4: 1B06 Light Chain
| Macromolecule | Name: 1B06 Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.77808 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVMTQSPAT LSVSPGDRAT LSCRASQSVS TELAWYQQKP GQAPRLLIYG ASTRATDIPA RFSGSGSGTE FTLTISSLQS EDFAVYFCQ QYNNWPPITF GQGTRLEIK |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Grid | Model: C-flat-1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | 1B06 in complex with a stabilized NC99 HA trimer |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 2.0 sec. / Average electron dose: 70.72 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: NOT APPLICABLE |
|---|---|
| Final 3D classification | Software - Name: cryoSPARC (ver. 2.15) |
| Final angle assignment | Type: NOT APPLICABLE |
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 3.96 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2.15) / Number images used: 73396 ) / Resolution.type: BY AUTHOR / Resolution: 3.96 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2.15) / Number images used: 73396 |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X










































