4FCY
 
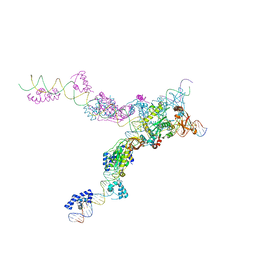 | | Crystal structure of the bacteriophage Mu transpososome | | Descriptor: | DNA (13-MER), DNA (49-MER), DNA (68-MER), ... | | Authors: | Montano, S.P, Pigli, Y.Z, Rice, P.A. | | Deposit date: | 2012-05-25 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.706 Å) | | Cite: | The Mu transpososome structure sheds light on DDE recombinase evolution.
Nature, 491, 2012
|
|
1M6U
 
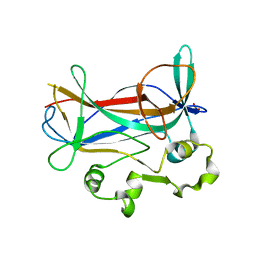 | | Crystal Structure of a Novel DNA-binding domain from Ndt80, a Transcriptional Activator Required for Meiosis in Yeast | | Descriptor: | Ndt80 protein, SULFATE ION | | Authors: | Montano, S.P, Cote, M.L, Fingerman, I, Pierce, M, Vershon, A.K, Georgiadis, M.M. | | Deposit date: | 2002-07-17 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M7U
 
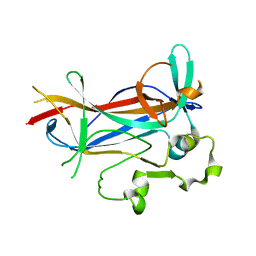 | | Crystal structure of a novel DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast | | Descriptor: | Ndt80 protein | | Authors: | Montano, S.P, Cote, M.L, Fingerman, I, Pierce, M, Vershon, A.K, Georgiadis, M.M. | | Deposit date: | 2002-07-22 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5CY1
 
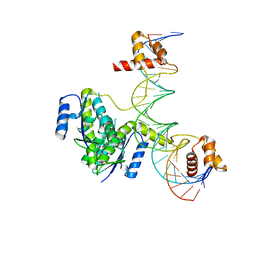 | |
2FVR
 
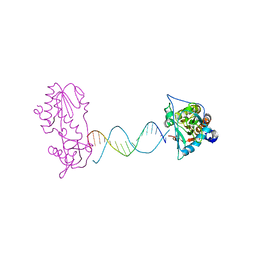 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*TP*CP*TP*TP*TP*CP*AP*TP*AP*TP*GP*AP*AP*AP*GP*A)-3', reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
2FVQ
 
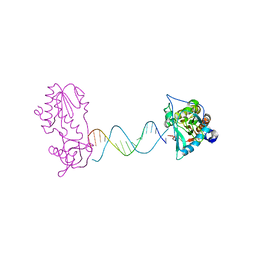 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*CP*TP*TP*TP*CP*AP*TP*TP*AP*AP*TP*GP*AP*AP*AP*G)-3', reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
2FVP
 
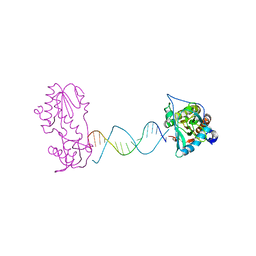 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*TP*TP*TP*CP*AP*TP*TP*GP*CP*AP*AP*TP*GP*AP*AP*A)-3', Reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
2FVS
 
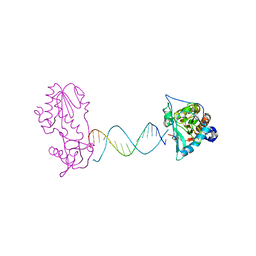 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*CP*AP*CP*AP*AP*TP*GP*AP*TP*CP*AP*TP*TP*GP*TP*G)-3', reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
4YHZ
 
 | | Crystal structure of 304M3-B Fab in complex with H3K4me3 peptide | | Descriptor: | Fab Heavy Chain, Fab Light Chain, GLYCEROL, ... | | Authors: | Hattori, T, Dementieva, I.S, Montano, S.P, Koide, S. | | Deposit date: | 2015-02-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YHY
 
 | | Crystal structure of 309M3-B in complex with trimethylated Lys | | Descriptor: | Fab Heavy Chain, Fab Light Chain, N-TRIMETHYLLYSINE | | Authors: | Hattori, T, Dementieva, I.S, Montano, S.P, Koide, S. | | Deposit date: | 2015-02-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YHP
 
 | | Crystal structure of 309M3-B Fab in complex with H3K9me3 peptide | | Descriptor: | Fab Heavy Chain, Fab Light Chain, H3K9me3 peptide | | Authors: | Hattori, T, Dementieva, I.S, Montano, S.P, Koide, S. | | Deposit date: | 2015-02-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1D0E
 
 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | 5'-D(*TP*TP*TP*CP*AP*TP*GP*CP*AP*TP*G)-3', REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M.L, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-09-09 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain
J.Mol.Biol., 296, 2000
|
|
1NND
 
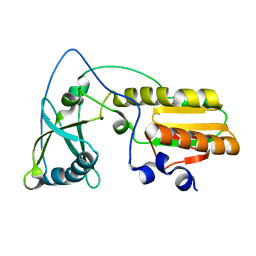 | | Arginine 116 is Essential for Nucleic Acid Recognition by the Fingers Domain of Moloney Murine Leukemia Virus Reverse Transcriptase | | Descriptor: | Reverse Transcriptase | | Authors: | Crowther, R.L, Remeta, D.P, Minetti, C.A, Das, D, Montano, S.P, Georgiadis, M.M. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and energetic characterization of nucleic acid-binding to the fingers domain of Moloney murine leukemia virus reverse transcriptase
Proteins, 57, 2004
|
|
1QAJ
 
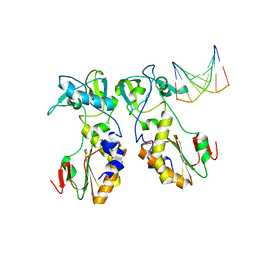 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-03-18 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
1QAI
 
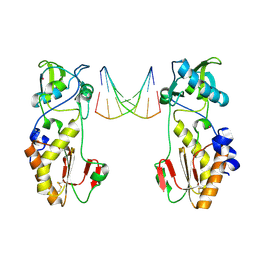 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), MERCURY (II) ION, REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
5CY2
 
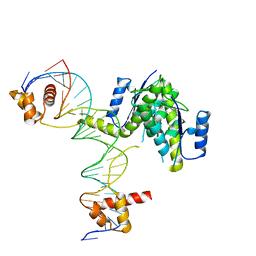 | |
1D1U
 
 | | USE OF AN N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE TO FACILITATE CRYSTALLIZATION AND ANALYSIS OF A PSEUDO-16-MER DNA MOLECULE CONTAINING G-A MISPAIRS | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*CP*AP*CP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*GP*TP*G)-3'), PROTEIN (REVERSE TRANSCRIPTASE) | | Authors: | Cote, M.L, Yohannan, S, Georgiadis, M.M. | | Deposit date: | 1999-09-21 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of an N-terminal fragment from moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing G-A mispairs.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1N4L
 
 | | A DNA analogue of the polypurine tract of HIV-1 | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*TP*AP*AP*AP*AP*AP*G)-3', 5'-D(*CP*TP*TP*TP*TP*TP*AP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', Reverse Transcriptase | | Authors: | Cote, M.L, Pflomm, M, Georgiadis, M.M. | | Deposit date: | 2002-10-31 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staying Straight with A-tracts: A DNA Analog of the HIV-1 Polypurine Tract
J.Mol.Biol., 330, 2003
|
|
