2ARW
 
 | | The solution structure of the membrane proximal cytokine receptor domain of the human interleukin-6 receptor | | Descriptor: | Interleukin-6 receptor alpha chain | | Authors: | Hecht, O, Dingley, A.J, Schwantner, A, Ozbek, S, Rose-John, S, Grotzinger, J. | | Deposit date: | 2005-08-22 | | Release date: | 2006-09-12 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the membrane-proximal cytokine receptor domain of the human interleukin-6 receptor
Biol.Chem., 387, 2006
|
|
1OF9
 
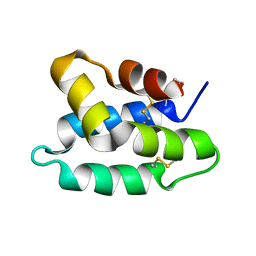 | | Solution structure of the pore forming toxin of entamoeba histolytica (Amoebapore A) | | Descriptor: | PORE-FORMING PEPTIDE AMEOBAPORE A | | Authors: | Hecht, O, Schleinkofer, K, Bruhn, H, Leippe, M, Van Nuland, N, Dingley, A.J, Grotzinger, J. | | Deposit date: | 2003-04-09 | | Release date: | 2004-02-26 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pore-Forming Protein of Entamoeba Histolytica
J.Biol.Chem., 279, 2004
|
|
2JSB
 
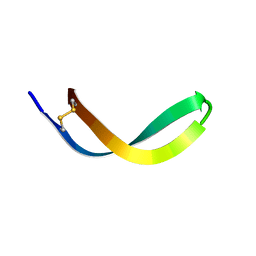 | | Solution structure of arenicin-1 | | Descriptor: | Arenicin-1 | | Authors: | Jakovkin, I.B, Hecht, O, Gelhaus, C, Krasnosdembskaya, A.D, Fedders, H, Leippe, M, Groetzinger, J. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-05 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structure and mode of action of the antimicrobial peptide arenicin
Biochem.J., 410, 2008
|
|
2W8B
 
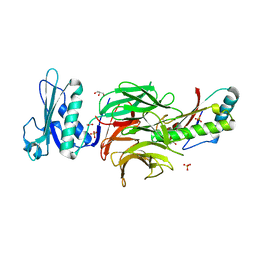 | | Crystal structure of processed TolB in complex with Pal | | Descriptor: | ACETATE ION, GLYCEROL, PEPTIDOGLYCAN-ASSOCIATED LIPOPROTEIN, ... | | Authors: | Sharma, A, Bonsor, D.A, Kleanthous, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Allosteric Beta-Propeller Signalling in Tolb and its Manipulation by Translocating Colicins.
Embo J., 28, 2009
|
|
3GHA
 
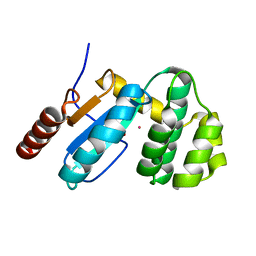 | | Crystal Structure of ETDA-treated BdbD (Reduced) | | Descriptor: | 1,2-ETHANEDIOL, Disulfide bond formation protein D, UNKNOWN ATOM OR ION | | Authors: | Crow, A, Lewin, A, Hederstedt, L, Le-Brun, N.E. | | Deposit date: | 2009-03-03 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure and Biophysical Properties of Bacillus subtilis BdbD: AN OXIDIZING THIOL:DISULFIDE OXIDOREDUCTASE CONTAINING A NOVEL METAL SITE
J.Biol.Chem., 284, 2009
|
|
3GH9
 
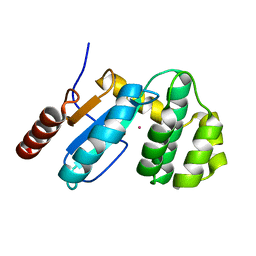 | | Crystal structure of EDTA-treated BdbD (Oxidised) | | Descriptor: | 1,2-ETHANEDIOL, Disulfide bond formation protein D, UNKNOWN ATOM OR ION | | Authors: | Crow, A, Lewin, A, Hederstedt, L, Le-Brun, N.E. | | Deposit date: | 2009-03-03 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure and Biophysical Properties of Bacillus subtilis BdbD: AN OXIDIZING THIOL:DISULFIDE OXIDOREDUCTASE CONTAINING A NOVEL METAL SITE
J.Biol.Chem., 284, 2009
|
|
6MRC
 
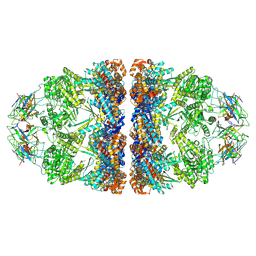 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
6MRD
 
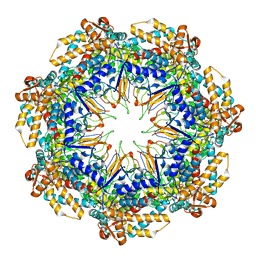 | | ADP-bound human mitochondrial Hsp60-Hsp10 half-football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
4DEX
 
 | |
4DEY
 
 | |
3BJ4
 
 | |
3CHM
 
 | |
3EU4
 
 | | Crystal Structure of BdbD from Bacillus subtilis (oxidised) | | Descriptor: | BdbD, CALCIUM ION | | Authors: | Crow, A, Moller, M.C, Hederstedt, L, Le Brun, N. | | Deposit date: | 2008-10-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Biophysical Properties of Bacillus subtilis BdbD: AN OXIDIZING THIOL:DISULFIDE OXIDOREDUCTASE CONTAINING A NOVEL METAL SITE
J.Biol.Chem., 284, 2009
|
|
3EU3
 
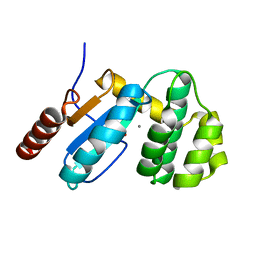 | | Crystal Structure of BdbD from Bacillus subtilis (reduced) | | Descriptor: | 1,2-ETHANEDIOL, BdbD, CALCIUM ION | | Authors: | Crow, A, Moller, M.C, Hederstedt, L, Le Brun, N.E. | | Deposit date: | 2008-10-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure and Biophysical Properties of Bacillus subtilis BdbD: AN OXIDIZING THIOL:DISULFIDE OXIDOREDUCTASE CONTAINING A NOVEL METAL SITE
J.Biol.Chem., 284, 2009
|
|
3QDP
 
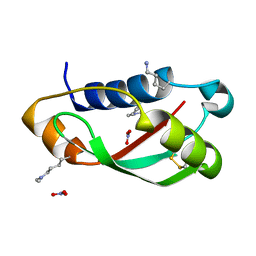 | |
3QDR
 
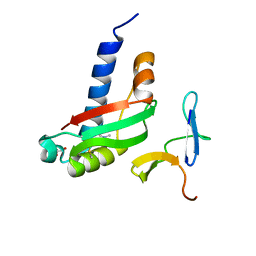 | |
2HT6
 
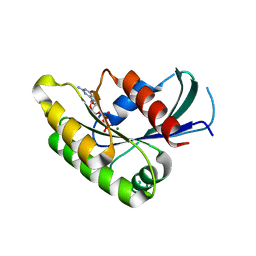 | |
2LLR
 
 | |
1T3S
 
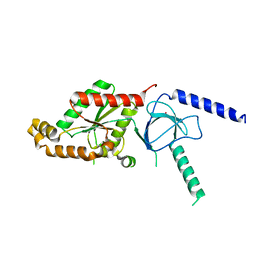 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, MERCURY (II) ION | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain.
Neuron, 42, 2004
|
|
1T3L
 
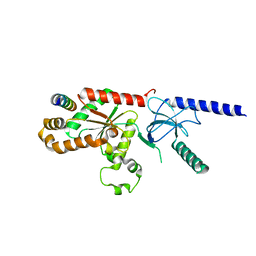 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core in Complex with Alpha1 Interaction Domain | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, Voltage-dependent L-type calcium channel alpha-1S subunit | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Voltage-Dependent Calcium Channel Beta Subunit Functional Core and Its Complex with the Alpha1 Interaction Domain
NEURON, 42, 2004
|
|
