8ORR
 
 | |
6GEW
 
 | | OphA Y63F-sinefungin complex | | Descriptor: | OphA, S-ADENOSYL-L-HOMOCYSTEINE, SINEFUNGIN | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2018-04-27 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
5OUF
 
 | |
6QZY
 
 | | full length OphA V406P in complex with SAH | | Descriptor: | ASN-GLY-PHE-PRO-TRP-MVA-ILE-MVA-VAL-GLY-PRO-ILE-GLY, MAGNESIUM ION, Peptide N-methyltransferase, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6QZZ
 
 | | full length OphA V404E in complex with SAH | | Descriptor: | Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6R00
 
 | | OphA DeltaC6 V404F complex with SAH | | Descriptor: | PHE-PRO-TRP-MVA-ILE-MVA-PHE-GLY-VAL-ILE-GLY-VAL-ILE-GLY, Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6TSC
 
 | | OphMA I407P complex with SAH | | Descriptor: | GLY-PHE-PRO-TRP-MVA-ILE-MVA-VAL-GLY-VAL-PRO-GLY, Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-12-20 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
1FQ1
 
 | | CRYSTAL STRUCTURE OF KINASE ASSOCIATED PHOSPHATASE (KAP) IN COMPLEX WITH PHOSPHO-CDK2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-DEPENDENT KINASE INHIBITOR 3, ... | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
1FPZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF KINASE ASSOCIATED PHOSPHATASE (KAP) WITH A SUBSTITUTION OF THE CATALYTIC SITE CYSTEINE (CYS140) TO A SERINE | | Descriptor: | CYCLIN-DEPENDENT KINASE INHIBITOR 3, SULFATE ION | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
6O5F
 
 | | Crystal structure of DEAD-box RNA helicase DDX3X at pre-unwound state | | Descriptor: | ATP-dependent RNA helicase DDX3X, CHLORIDE ION, RNA (5'-R(P*CP*AP*AP*GP*GP*UP*CP*AP*UP*UP*CP*GP*CP*AP*AP*GP*AP*GP*UP*GP*GP*CP*C)-3') | | Authors: | Song, H, Ji, X. | | Deposit date: | 2019-03-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X.
Nat Commun, 10, 2019
|
|
6PC1
 
 | | Crystal structure of Helicobacter pylori PPX/GppA (E143A) in complex with ppGpp | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PC0
 
 | | Crystal structure of Helicobacter pylori PPX/GppA | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, D-MALATE, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PBZ
 
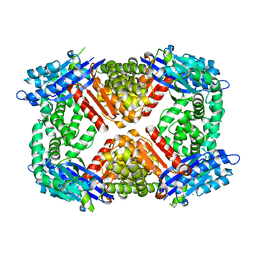 | | Crystal structure of Escherichia coli GppA | | Descriptor: | CHLORIDE ION, Guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase | | Authors: | Song, H, Shaw, G.X, Wang, C, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.475 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PC3
 
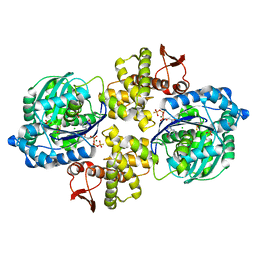 | | Crystal structure of Helicobacter pylori PPX/GppA in complex with GSP | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PC2
 
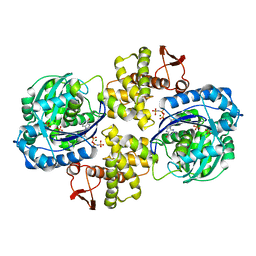 | | Crystal structure of Helicobacter pylori PPX/GppA in complex with GNP | | Descriptor: | Guanosine pentaphosphate phosphohydrolase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
1EFC
 
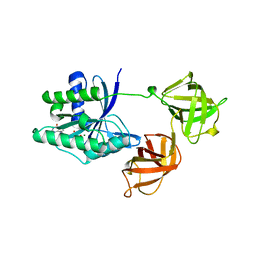 | | INTACT ELONGATION FACTOR FROM E.COLI | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (ELONGATION FACTOR) | | Authors: | Song, H, Parsons, M.R, Rowsell, S, Leonard, G, Phillips, S.E.V. | | Deposit date: | 1998-11-24 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution.
J.Mol.Biol., 285, 1999
|
|
5N0V
 
 | |
5N0W
 
 | |
5N0N
 
 | |
5N0U
 
 | |
5N0X
 
 | | Crystal structure of OphA-DeltaC6 in complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Peptide N-Methyltransferase, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
5N0T
 
 | |
5N0S
 
 | |
5N0R
 
 | |
5N4I
 
 | |
