2C61
 
 | | Crystal structure of the non-catalytic B subunit of A-type ATPase from M. mazei Go1 | | Descriptor: | A-TYPE ATP SYNTHASE NON-CATALYTIC SUBUNIT B | | Authors: | Schaefer, I, Bailer, S.M, Dueser, M, Boersch, M, Bernal, R.A, Grueber, G, Stock, D. | | Deposit date: | 2005-11-06 | | Release date: | 2006-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Archaeal A1Ao ATP Synthase Subunit B from Methanosarcina Mazei Go1: Implications of Nucleotide-Binding Differences in the Major A1Ao Subunits a and B. Subunits a and B
J.Mol.Biol., 358, 2006
|
|
3SSA
 
 | | Crystal structure of subunit B mutant N157T of the A1AO ATP synthase | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sundararaman, L, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-07-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
3TIV
 
 | | Crystal structure of subunit B mutant N157A of the A1AO ATP synthase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-08-22 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
3TGW
 
 | | Crystal structure of subunit B mutant H156A of the A1AO ATP synthase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-08-18 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
2JDI
 
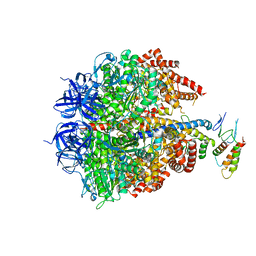 | | Ground state structure of F1-ATPase from bovine heart mitochondria (Bovine F1-ATPase crystallised in the absence of azide) | | Descriptor: | ATP SYNTHASE DELTA CHAIN, ATP SYNTHASE EPSILON CHAIN, ATP SYNTHASE GAMMA CHAIN, ... | | Authors: | Bowler, M.W, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ground State Structure of F1-ATPase from Bovine Heart Mitochondria at 1.9 A Resolution
J.Biol.Chem., 282, 2007
|
|
2CK3
 
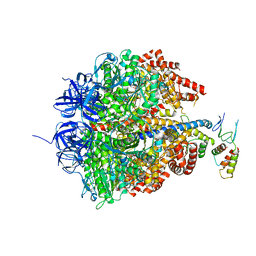 | | Azide inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA, MITOCHONDRIAL, ... | | Authors: | Bowler, M.W, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How Azide Inhibits ATP Hydrolysis by the F-Atpases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1H8E
 
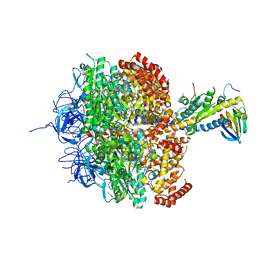 | | (ADP.AlF4)2(ADP.SO4) bovine F1-ATPase (all three catalytic sites occupied) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, GLYCEROL, ... | | Authors: | Menz, R.I, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2001-02-02 | | Release date: | 2001-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase with Nucleotide Bound to All Three Catalytic Sites: Implications for the Mechanism of Rotary Catalysis
Cell(Cambridge,Mass.), 106, 2001
|
|
5BN3
 
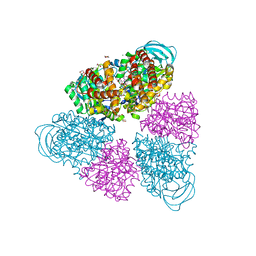 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ADP from Nanoarcheaum equitans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
2R9V
 
 | |
2V7Q
 
 | | The structure of F1-ATPase inhibited by I1-60HIS, a monomeric form of the inhibitor protein, IF1. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP SYNTHASE DELTA CHAIN, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How the Regulatory Protein, If1, Inhibits F1- ATPase from Bovine Mitochondria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3B2Q
 
 | | Intermediate position of ATP on its trail to the binding pocket inside the subunit B mutant R416W of the energy converter A1Ao ATP synthase | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, ... | | Authors: | Kumar, A, Manimekalai, M.S.S, Balakrishna, A.M, Hunke, C, Gruber, G. | | Deposit date: | 2007-10-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Spectroscopic and crystallographic studies of the mutant R416W give insight into the nucleotide binding traits of subunit B of the A1Ao ATP synthase
Proteins, 75, 2009
|
|
5ZE9
 
 | | Crystal structure of AMP-PNP bound mutant A3B3 complex from Enterococcus hirae V-ATPase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maruyama, S, Suzuki, K, Sasaki, H, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
7NK7
 
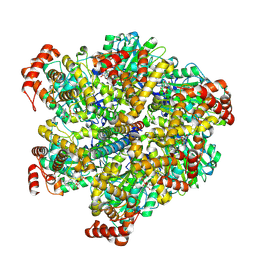 | | Mycobacterium smegmatis ATP synthase F1 state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-17 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NKJ
 
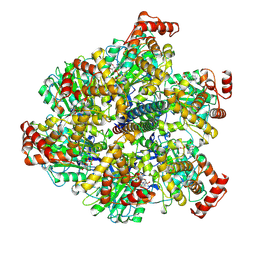 | | Mycobacterium smegmatis ATP synthase F1 state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3VR4
 
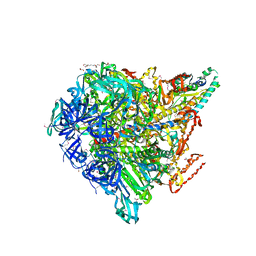 | | Crystal structure of Enterococcus hirae V1-ATPase [eV1] | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
4Q4L
 
 | |
1W0J
 
 | | Beryllium fluoride inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, MITOCHONDRIAL PRECURSOR, ... | | Authors: | Kagawa, R, Montgomery, M.G, Braig, K, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Bovine F1-ATPase Inhibited by Adp and Beryllium Fluoride
Embo J., 23, 2004
|
|
5CDF
 
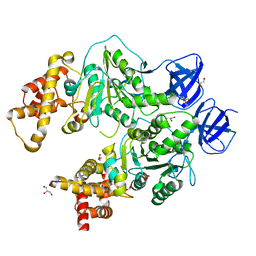 | | Structure at 2.3 A of the alpha/beta monomer of the F-ATPase from Paracoccus denitrificans | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, GLYCEROL, ... | | Authors: | Morales-Rios, E, Montgomery, M.G, Leslie, A.G.W, Garcia-Trejo, J.J, Walker, J.E. | | Deposit date: | 2015-07-03 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a catalytic dimer of the alpha- and beta-subunits of the F-ATPase from Paracoccus denitrificans at 2.3 angstrom resolution.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2JIZ
 
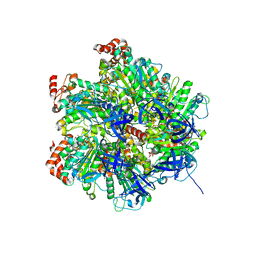 | | The Structure of F1-ATPase inhibited by resveratrol. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3MFY
 
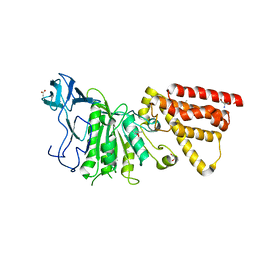 | | Structural characterization of the subunit A mutant F236A of the A-ATP synthase from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Balakrishna, A.M, Kumar, A, Manimekali, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-04-05 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
3QJY
 
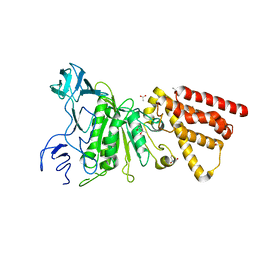 | | Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3M4Y
 
 | | Structural characterization of the subunit A mutant P235A of the A-ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Manimekalai, M.S, Balakrishna, A.M, Kumar, A, Priya, R, Biukovic, G, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-03-12 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
3I73
 
 | | Structural characterization for the nucleotide binding ability of subunit A with ADP of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A-TYPE ATP SYNTHASE CATALYTIC SUBUNIT A, ... | | Authors: | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2009-07-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
3IKJ
 
 | | Structural characterization for the nucleotide binding ability of subunit A mutant S238A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, V-type ATP synthase alpha chain | | Authors: | Kumar, A, Manimekali, M.S.S, Balakrishna, A.M, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2009-08-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
3ND8
 
 | | Structural characterization for the nucleotide binding ability of subunit A of the A1AO ATP synthase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | Authors: | Kumar, A, Gruber, G. | | Deposit date: | 2010-06-07 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The transition-like state and Pi entrance into the catalytic a subunit of the biological engine A-ATP synthase.
J.Mol.Biol., 408, 2011
|
|
