6G2U
 
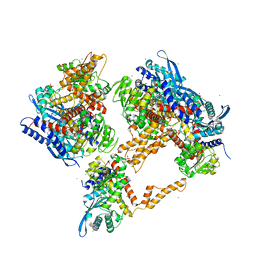 | | Crystal structure of the human glutamate dehydrogenase 2 (hGDH2) | | Descriptor: | CHLORIDE ION, Glutamate dehydrogenase 2, mitochondrial, ... | | Authors: | Fadouloglou, V.F, Dimovasili, C, Providaki, M, Kotsifaki, D, Sarrou, I, Plaitakis, A, Zaganas, I, Kokkinidis, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.934287 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 2, a positively selected novel human enzyme involved in brain biology and cancer pathophysiology.
J.Neurochem., 2021
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3K8Z
 
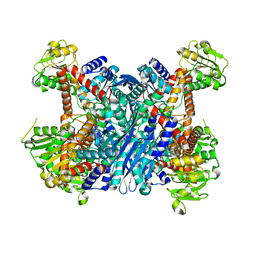 | | Crystal Structure of Gudb1 a decryptified secondary glutamate dehydrogenase from B. subtilis | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Gunka, K, Newman, J.A, Commichau, F.M, Herzberg, C, Rodrigues, C, Hewitt, L, Lewis, R.J, Stulke, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional dissection of a trigger enzyme: mutations of the bacillus subtilis glutamate dehydrogenase RocG that affect differentially its catalytic activity and regulatory properties
J.Mol.Biol., 400, 2010
|
|
3JCZ
 
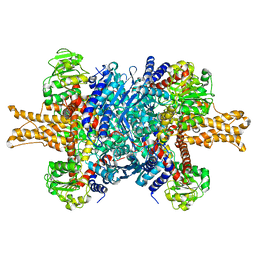 | | Structure of bovine glutamate dehydrogenase in the unliganded state | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD3
 
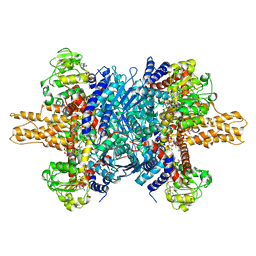 | | Glutamate dehydrogenase in complex with NADH and GTP, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD2
 
 | | Glutamate dehydrogenase in complex with NADH, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
1B3B
 
 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT N97D, G376K | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Knapp, S, Lebbink, J.H.G, Van Der Oost, J, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-07 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. I. Introduction of a six-residue ion-pair network in the hinge region.
J.Mol.Biol., 280, 1998
|
|
1AUP
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | Authors: | Baker, P.J, Waugh, M.L, Stillman, T.J, Turnbull, A.P, Rice, D.W. | | Deposit date: | 1997-09-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determinants of substrate specificity in the superfamily of amino acid dehydrogenases.
Biochemistry, 36, 1997
|
|
1B26
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Knapp, S, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glutamate dehydrogenase from the hyperthermophilic eubacterium Thermotoga maritima at 3.0 A resolution.
J.Mol.Biol., 267, 1997
|
|
1BGV
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, GLUTAMIC ACID | | Authors: | Stillman, T.J, Baker, P.J, Britton, K.L, Rice, D.W. | | Deposit date: | 1998-06-01 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis.
J.Mol.Biol., 234, 1993
|
|
1BXG
 
 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND BETA-PHENYLPROPIONATE | | Descriptor: | HYDROCINNAMIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-02 | | Release date: | 1999-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
1C1X
 
 | | L-PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND L-3-PHENYLLACTATE | | Descriptor: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, ISOPROPYL ALCOHOL, L-PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B. | | Deposit date: | 1999-07-22 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
Biochemistry, 39, 2000
|
|
1BVU
 
 | | GLUTAMATE DEHYDROGENASE FROM THERMOCOCCUS LITORALIS | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Baker, P.J, Britton, K.L, Yip, K.S, Stillman, T.J, Rice, D.W. | | Deposit date: | 1999-07-20 | | Release date: | 1999-09-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of the glutamate dehydrogenase from the hyperthermophile Thermococcus litoralis and its comparison with that from Pyrococcus furiosus
J.Mol.Biol., 293, 1999
|
|
1BW9
 
 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND PHENYLPYRUVATE | | Descriptor: | 1,2-ETHANEDIOL, 3-PHENYLPYRUVIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
1C1D
 
 | | L-PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NADH AND L-PHENYLALANINE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ISOPROPYL ALCOHOL, L-PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B. | | Deposit date: | 1999-07-21 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
Biochemistry, 39, 2000
|
|
8XCU
 
 | |
8XD3
 
 | |
8XCO
 
 | |
8XCW
 
 | |
8XCZ
 
 | |
8XD0
 
 | |
8XD5
 
 | |
8XD6
 
 | |
8XCS
 
 | |
8XCX
 
 | |
