7K06
 
 | |
7LPY
 
 | |
7LPZ
 
 | |
7LQ0
 
 | |
2F96
 
 | | 2.1 A crystal structure of Pseudomonas aeruginosa rnase T (Ribonuclease T) | | Descriptor: | MAGNESIUM ION, Ribonuclease T | | Authors: | Zheng, H, Chruszcz, M, Cymborowski, M, Wang, Y, Gorodichtchenskaia, E, Skarina, T, Guthrie, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of RNase T, an Exoribonuclease Involved in tRNA Maturation and End Turnover.
Structure, 15, 2007
|
|
4FZX
 
 | | Exonuclease X in complex with 3' overhanging duplex DNA | | Descriptor: | DNA (5'-D(P*CP*GP*GP*AP*TP*CP*CP*AP*CP*AP*AP*TP*GP*AP*CP*CP*T)-3'), DNA (5'-D(P*GP*TP*CP*AP*TP*TP*GP*TP*GP*GP*AP*TP*CP*CP*GP*AP*G)-3'), Exodeoxyribonuclease 10, ... | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
4FZY
 
 | | Exonuclease X in complex with 12bp blunt-ended dsDNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*GP*AP*TP*TP*CP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*GP*AP*AP*TP*CP*TP*AP*CP*A)-3'), Exodeoxyribonuclease 10, ... | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
4FZZ
 
 | | Exonuclease X in complex with 5' overhanging duplex DNA | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*TP*GP*TP*GP*GP*AP*TP*CP*CP*GP*AP*G)-3'), Exodeoxyribonuclease 10, SODIUM ION | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
2GBZ
 
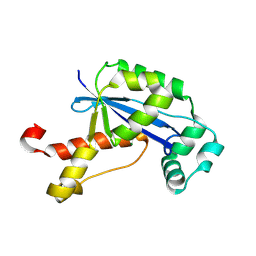 | | The Crystal Structure of XC847 from Xanthomonas campestris: a 3-5 Oligoribonuclease of DnaQ fold family with a Novel Opposingly-Shifted Helix | | Descriptor: | MAGNESIUM ION, Oligoribonuclease | | Authors: | Chin, K.H, Yang, C.Y, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-03-12 | | Release date: | 2007-01-16 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of XC847 from Xanthomonas campestris: a 3'-5' oligoribonuclease of DnaQ fold family with a novel opposingly shifted helix
Proteins, 65, 2006
|
|
2GUI
 
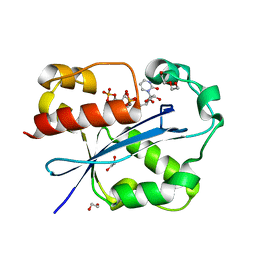 | | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit of E. coli DNA Polymerase III | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA polymerase III epsilon subunit, ... | | Authors: | Park, A.Y, Carr, P.D, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2006-04-30 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit E. coli DNA Polymerase III
To be Published
|
|
3CG7
 
 | |
3CM6
 
 | |
3CM5
 
 | |
2IGI
 
 | | Crystal Structure of E. coli Oligoribonuclease | | Descriptor: | ACETIC ACID, CADMIUM ION, Oligoribonuclease, ... | | Authors: | Zuo, Y, Malhotra, A. | | Deposit date: | 2006-09-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Oligoribonuclease, the lone essential exoribonuclease in Escherichia coli
To be Published
|
|
2IS3
 
 | |
3V9W
 
 | |
3V9Z
 
 | |
3V9S
 
 | |
3V9U
 
 | |
3VA3
 
 | |
3V9X
 
 | |
3VA0
 
 | |
7V5H
 
 | |
7VA6
 
 | |
7VA2
 
 | |
