1FN0
 
 | | STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14D. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
3ZC9
 
 | | Crystal Structure of Murraya koenigii Miraculin-Like Protein at 2.2 A resolution at pH 4.6 | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Selvakumar, P, Sharma, N, Tomar, P.P.S, Kumar, P, Sharma, A.K. | | Deposit date: | 2012-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights Into the Aggregation Behavior of Murraya Koenigii Miraculin-Like Protein Below Ph 7.5.
Proteins, 82, 2014
|
|
2WBC
 
 | | REFINED CRYSTAL STRUCTURE (2.3 ANGSTROM) OF A WINGED BEAN CHYMOTRYPSIN INHIBITOR AND LOCATION OF ITS SECOND REACTIVE SITE | | Descriptor: | CHYMOTRYPSIN INHIBITOR | | Authors: | Dattagupta, J.K, Podder, A, Chakrabarti, C, Sen, U, Mukhopadhyay, D, Dutta, S.K, Singh, M. | | Deposit date: | 1997-11-26 | | Release date: | 1998-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined crystal structure (2.3 A) of a double-headed winged bean alpha-chymotrypsin inhibitor and location of its second reactive site.
Proteins, 35, 1999
|
|
3S8J
 
 | |
3S8K
 
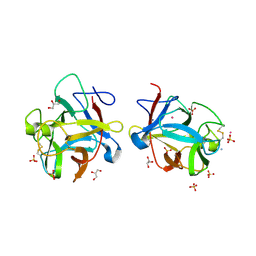 | |
2QN4
 
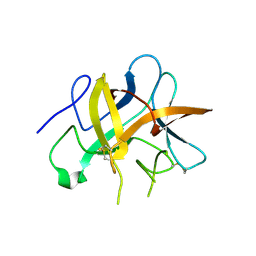 | | Structure and function study of rice bifunctional alpha-amylase/subtilisin inhibitor from Oryza sativa | | Descriptor: | Alpha-amylase/subtilisin inhibitor | | Authors: | Peng, W.Y, Lin, Y.H, Huang, Y.C, Guan, H.H, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function Study of Rice Bifunctional Alpha-Amylase/Subtilisin Inhibitor from Oryza Sativa
To be Published
|
|
3ZC8
 
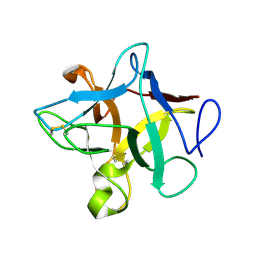 | | Crystal Structure of Murraya koenigii Miraculin-Like Protein at 2.2 A resolution at pH 7.0 | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Selvakumar, P, Sharma, N, Tomar, P.P.S, Kumar, P, Sharma, A.K. | | Deposit date: | 2012-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights Into the Aggregation Behavior of Murraya Koenigii Miraculin-Like Protein Below Ph 7.5.
Proteins, 82, 2014
|
|
1EYL
 
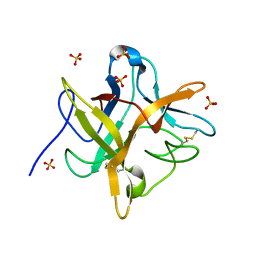 | | STRUCTURE OF A RECOMBINANT WINGED BEAN CHYMOTRYPSIN INHIBITOR | | Descriptor: | CHYMOTRYPSIN INHIBITOR, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Ghosh, S. | | Deposit date: | 2000-05-07 | | Release date: | 2000-05-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
Protein Eng., 14, 2001
|
|
2DRE
 
 | | Crystal structure of Water-soluble chlorophyll protein from lepidium virginicum at 2.00 angstrom resolution | | Descriptor: | CHLOROPHYLL A, Water-soluble chlorophyll protein | | Authors: | Horigome, D, Satoh, H, Itoh, N, Mitsunaga, K, Oonishi, I, Nakagawa, A, Uchida, A. | | Deposit date: | 2006-06-08 | | Release date: | 2006-12-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism and photoprotective function of water-soluble chlorophyll-binding protein.
J.Biol.Chem., 282, 2007
|
|
4II0
 
 | |
4IHZ
 
 | |
4J2K
 
 | | Crystal structure of a plant trypsin inhibitor EcTI | | Descriptor: | GLYCEROL, IMIDAZOLE, Trypsin inhibitor | | Authors: | Zhou, D, Wlodawer, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of a Plant Trypsin Inhibitor from Enterolobium contortisiliquum (EcTI) and of Its Complex with Bovine Trypsin.
Plos One, 8, 2013
|
|
2GZB
 
 | | Bauhinia bauhinioides cruzipain inhibitor (BbCI) | | Descriptor: | IODIDE ION, Kunitz-type proteinase inhibitor BbCI | | Authors: | Hansen, D, Macedo-Ribeiro, S, Navarro, M.V.A.S, Garratt, R.C, Oliva, M.L.V. | | Deposit date: | 2006-05-11 | | Release date: | 2007-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a novel cysteinless plant Kunitz-type protease inhibitor.
Biochem.Biophys.Res.Commun., 360, 2007
|
|
5XOZ
 
 | |
3I2A
 
 | |
3I2X
 
 | | Crystal structure of a chimeric trypsin inhibitor having reactive site loop of ETI on the scaffold of WCI | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Sen, U, Khamrui, S, Dasgupta, J, Dattagupta, J.K, Majumder, S. | | Deposit date: | 2009-06-30 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of a novel set of scaffolding residues that are instrumental for the inhibitory property of Kunitz (STI) inhibitors.
Protein Sci., 19, 2010
|
|
3IIR
 
 | | Crystal Structure of Miraculin like protein from seeds of Murraya koenigii | | Descriptor: | Trypsin inhibitor | | Authors: | Gahloth, D, Selvakumar, P, Shee, C, Kumar, P, Sharma, A.K. | | Deposit date: | 2009-08-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cloning, sequence analysis and crystal structure determination of a miraculin-like protein from Murraya koenigii
Arch.Biochem.Biophys., 494, 2010
|
|
5WVX
 
 | | Crystal Structure of bifunctional Kunitz type Trypsin /amylase inhibitor (AMTIN) from the tubers of Alocasia macrorrhiza | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CITRIC ACID, Trypsin/chymotrypsin inhibitor | | Authors: | Palayam, M, Radhakrishnan, M, Lakshminarayanan, K, Balu, K.E, Ganapathy, J, Krishnasamy, G. | | Deposit date: | 2016-12-29 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural insights into a multifunctional inhibitor, 'AMTIN' from tubers of Alocasia macrorrhizos and its possible role in dengue protease (NS2B-NS3) inhibition.
Int. J. Biol. Macromol., 113, 2018
|
|
2GO2
 
 | |
5YCZ
 
 | | Crystal structure of Alocasin, protease inhibitor from Giant Taro (Arum macrorrhizon) | | Descriptor: | Trypsin/chymotrypsin inhibitor | | Authors: | Vajravijayan, S, Pletnev, S, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2017-09-08 | | Release date: | 2018-06-13 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structure of a novel Kunitz type inhibitor, alocasin with anti-Aedes aegypti activity targeting midgut proteases.
Pest Manag. Sci., 74, 2018
|
|
5YH4
 
 | | Miraculin-like protein from Vitis vinifera | | Descriptor: | 1,2-ETHANEDIOL, mirauclin-like protein | | Authors: | Shimizu-Ibuka, A, Furukawa, N. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and functional analysis of miraculin-like protein from Vitis vinifera.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
2ESU
 
 | | Crystal structure of Asn to Gln mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
1BA7
 
 | | SOYBEAN TRYPSIN INHIBITOR | | Descriptor: | TRYPSIN INHIBITOR (KUNITZ) | | Authors: | De Meester, P, Brick, P, Lloyd, L.F, Blow, D.M, Onesti, S. | | Deposit date: | 1998-04-22 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Kunitz-type soybean trypsin inhibitor (STI): implication for the interactions between members of the STI family and tissue-plasminogen activator.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AVU
 
 | | TRYPSIN INHIBITOR FROM SOYBEAN (STI) | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-20 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
5DSS
 
 | |
