7GRF
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-2 | | 分子名称: | 3C-like proteinase nsp5, 5-bromopyridin-3-amine, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS4
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-27 | | 分子名称: | 3C-like proteinase nsp5, 7-(hydroxymethyl)-3-methyl-6~{H}-[1,3]thiazolo[3,2-a]pyrimidin-5-one, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRU
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-17 | | 分子名称: | 3-(4-chlorophenyl)-1-methyl-1H-pyrazol-5-amine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS3
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-26 | | 分子名称: | (6-phenylpyridin-3-yl)methanamine, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRK
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-7 | | 分子名称: | (6-fluoro-2H,4H-1,3-benzodioxin-8-yl)methanol, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRS
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-15 | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, SODIUM ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS0
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-23 | | 分子名称: | (pyridin-2-yl)(quinolin-2-yl)methanone, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRP
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-12 | | 分子名称: | 1-(2,3-dihydro-1-benzofuran-5-yl)methanamine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRW
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-19 | | 分子名称: | (2S)-N-(3,5-dichlorophenyl)-2-hydroxypropanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8X3H
 
 | |
8R43
 
 | |
8UXX
 
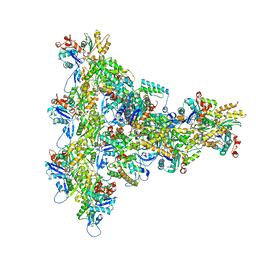 | | Arp2/3 branch junction complex, BeFx state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | 登録日 | 2023-11-11 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8R3V
 
 | |
8UXM
 
 | |
8UXL
 
 | |
8R37
 
 | | Klebsiella pneumoniae fosfomycin-resistance protein (FosAKP) | | 分子名称: | FOSFOMYCIN, FosA family fosfomycin resistance glutathione transferase, L(+)-TARTARIC ACID, ... | | 著者 | Papageorgiou, A.C, Varotsou, C, Labrou, N.E. | | 登録日 | 2023-11-08 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Structural Studies of Klebsiella pneumoniae Fosfomycin-Resistance Protein and Its Application for the Development of an Optical Biosensor for Fosfomycin Determination.
Int J Mol Sci, 25, 2023
|
|
8R34
 
 | | CryoEM structure of the symmetric Pho90 dimer from yeast with substrates. | | 分子名称: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Low-affinity phosphate transporter PHO90, PHOSPHATE ION, ... | | 著者 | Schneider, S, Kuehlbrandt, W, Yildiz, O. | | 登録日 | 2023-11-08 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Complementary structures of the yeast phosphate transporter Pho90 provide insights into its transport mechanism.
Structure, 2024
|
|
8X1H
 
 | |
8R2I
 
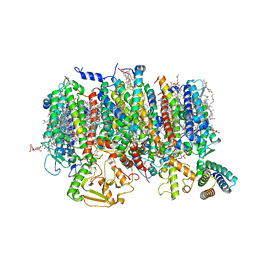 | | Cryo-EM Structure of native Photosystem II assembly intermediate from Chlamydomonas reinhardtii | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | 著者 | Fadeeva, M, Klaiman, D, Kandiah, E, Nelson, N. | | 登録日 | 2023-11-06 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of native photosystem II assembly intermediate from Chlamydomonas reinhardtii .
Front Plant Sci, 14, 2023
|
|
8R16
 
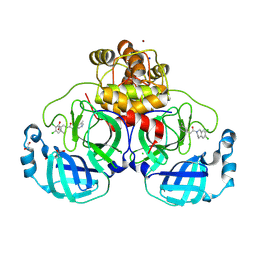 | | Structure of compound 12 bound to SARS-CoV-2 main protease | | 分子名称: | 1,2-ETHANEDIOL, 1-[6,7-bis(chloranyl)-3,4-dihydro-1H-isoquinolin-2-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R12
 
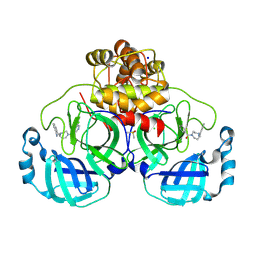 | | Structure of compound 8 bound to SARS-CoV-2 main protease | | 分子名称: | 2-[[4-(5-chloranylpyridin-3-yl)carbonyl-1,4-diazepan-1-yl]methyl]benzenecarbonitrile, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.587 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R11
 
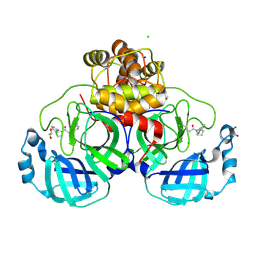 | | Structure of compound 7 bound to SARS-CoV-2 main protease | | 分子名称: | 1,2-ETHANEDIOL, 1-[(2~{S})-2-(3-chlorophenyl)pyrrolidin-1-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R14
 
 | | Structure of compound 11 bound to SARS-CoV-2 main protease | | 分子名称: | (5-chloranylpyridin-3-yl)-[4-[(2-chlorophenyl)methyl]-1,4-diazepan-1-yl]methanone, 3C-like proteinase, BROMIDE ION, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.336 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R1D
 
 | | SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-3 Fab Heavy Chain, SD1-3 Fab Light Chain, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | 登録日 | 2023-11-01 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.37 Å) | | 主引用文献 | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8UU5
 
 | | Cryo-EM structure of the Listeria innocua 70S ribosome (head-swiveled) in complex with pe/E-tRNA (structure I-B) | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | 著者 | Seely, S.M, Basu, R.S, Gagnon, M.G. | | 登録日 | 2023-10-31 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
