3L6W
 
 | | Structure of the collar functional unit (KLH1-H) of keyhole limpet hemocyanin | | Descriptor: | Hemocyanin 1 | | Authors: | Jaenicke, E, Buechler, K, Markl, J, Decker, H, Barends, T.R.M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Cupredoxin-like Domains in Hemocyanins.
Biochem.J., 2009
|
|
4Z12
 
 | | Recombinantly expressed latent aurone synthase (polyphenol oxidase) co-crystallized with hexatungstotellurate(VI) | | Descriptor: | 6-tungstotellurate(VI), Aurone synthase, COPPER (II) ION, ... | | Authors: | Molitor, C, Mauracher, S.G, Rompel, A. | | Deposit date: | 2015-03-26 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Aurone synthase is a catechol oxidase with hydroxylase activity and provides insights into the mechanism of plant polyphenol oxidases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Z13
 
 | | Recombinantly expressed latent aurone synthase (polyphenol oxidase) co-crystallized with hexatungstotellurate(VI) and soaked in H2O2 | | Descriptor: | 6-tungstotellurate(VI), Aurone synthase, COPPER (II) ION, ... | | Authors: | Molitor, C, Mauracher, S.G, Rompel, A. | | Deposit date: | 2015-03-26 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Aurone synthase is a catechol oxidase with hydroxylase activity and provides insights into the mechanism of plant polyphenol oxidases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Z10
 
 | | Inactive aurone synthase (polyphenol oxidase) co-crystallized with 1,4-resorcinol | | Descriptor: | ACETATE ION, Aurone synthase, COPPER (II) ION, ... | | Authors: | Molitor, C, Mauracher, S.G, Rompel, A. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inactive aurone synthase (polyphenol oxidase) co-crystallized with 1,4-resorcinol
To Be Published
|
|
4Z0Z
 
 | |
4Z11
 
 | |
4Z0Y
 
 | | Active aurone synthase (polyphenol oxidase), copper B : sulfohistidine ~ 1.4 : 1 | | Descriptor: | Aurone synthase, COPPER (II) ION, GLYCEROL | | Authors: | Molitor, C, Mauracher, S.G, Rompel, A. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aurone synthase is a catechol oxidase with hydroxylase activity and provides insights into the mechanism of plant polyphenol oxidases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Z14
 
 | |
6J2U
 
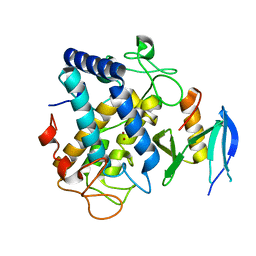 | |
6HQJ
 
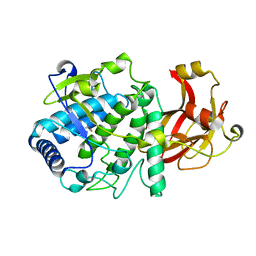 | |
6HQI
 
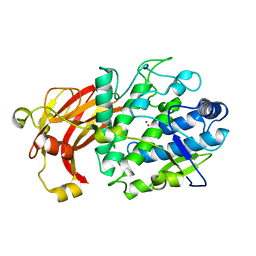 | | holo-form of polyphenol oxidase from Solanum lycopersicum | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Polyphenol oxidase A, ... | | Authors: | Kampatsikas, I, Bijelic, A, Rompel, A. | | Deposit date: | 2018-09-25 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of tomato polyphenol oxidases provide novel insights into their substrate specificity.
Sci Rep, 9, 2019
|
|
6JUD
 
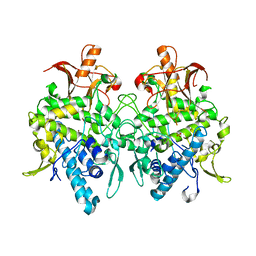 | | Radiation damage in Aspergillus oryzae pro-tyrosinase oxygen-bound C92A/H103F mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5CE9
 
 | | structure of tyrosinase from walnut (Juglans regia) | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Polyphenol oxidase, ... | | Authors: | Bijelic, A, Pretzler, M, Zekiri, F, Rompel, A. | | Deposit date: | 2015-07-06 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of a Plant Tyrosinase from Walnut Leaves Reveals the Importance of """"Substrate-Guiding Residues"""" for Enzymatic Specificity.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6JUC
 
 | | Aspergillus oryzae pro-tyrosinase oxygen-bound C92A/H103F mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU5
 
 | | Aspergillus oryzae pro-tyrosinase C92A/F513Y mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5Z0E
 
 | |
5Z0L
 
 | |
5Z0F
 
 | |
5Z0M
 
 | |
5Z0K
 
 | |
5Z0D
 
 | |
5Z0J
 
 | |
5Z0H
 
 | |
5Z0I
 
 | |
5M6B
 
 | |
