1SQE
 
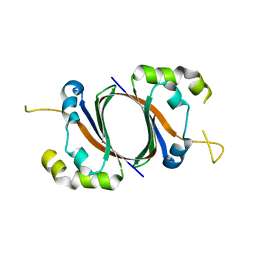 | | 1.5A Crystal Structure Of the protein PG130 from Staphylococcus aureus, Structural genomics | | Descriptor: | hypothetical protein PG130 | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases
J.Biol.Chem., 280, 2005
|
|
1R6Y
 
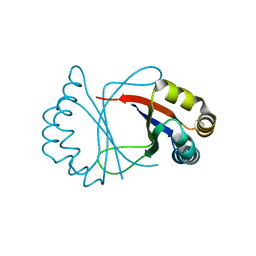 | |
3HX9
 
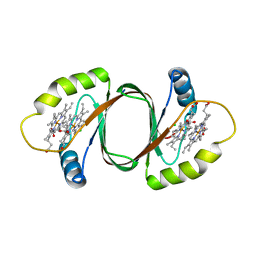 | | Structure of heme-degrader, MhuD (Rv3592), from Mycobacterium tuberculosis with two hemes bound in its active site | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Protein Rv3592 | | Authors: | Chim, N, Nguyen, T.Q, Iniguez, A, Goulding, C.W. | | Deposit date: | 2009-06-19 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unusual Diheme Conformation of the Heme-Degrading Protein from Mycobacterium tuberculosis
J.Mol.Biol., 395, 2009
|
|
1TZ0
 
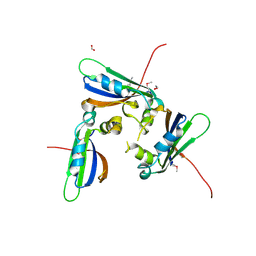 | |
1TUV
 
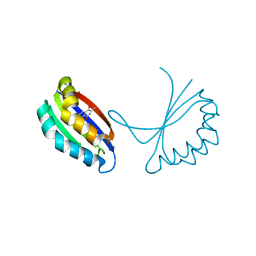 | | Crystal structure of YgiN in complex with menadione | | Descriptor: | MENADIONE, Protein ygiN | | Authors: | Adams, M.A, Jia, Z. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Evidence for an Enzymatic Quinone Redox Cycle in Escherichia coli: IDENTIFICATION OF A NOVEL QUINOL MONOOXYGENASE
J.Biol.Chem., 280, 2005
|
|
3GZ7
 
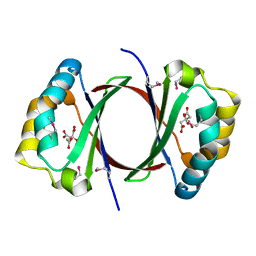 | |
1Q8B
 
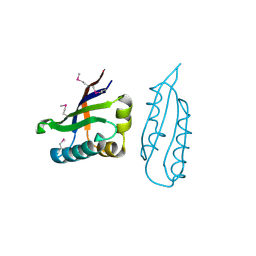 | |
3KG1
 
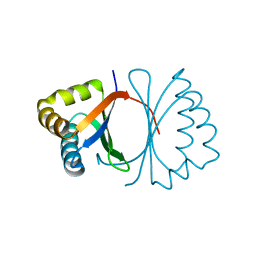 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, mutant N63A | | Descriptor: | CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KG0
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.7 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KKF
 
 | |
