9IMJ
 
 | |
9GGQ
 
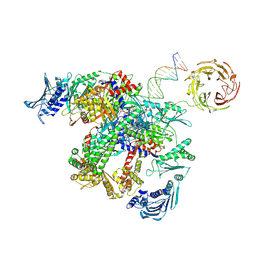 | | E.coli gyrase holocomplex with cleaved chirally wrapped 217 bp DNA fragment and moxifloxacin | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Ghilarov, D, Heddle, J.G, Pabis, M. | | Deposit date: | 2024-08-13 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of chiral wrap and T-segment capture by Escherichia coli DNA gyrase
Proceedings of the National Academy of Sciences USA, 2024
|
|
9GBV
 
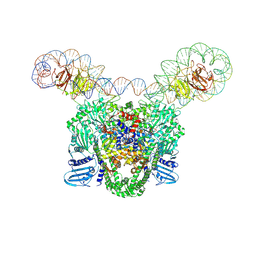 | |
9EUY
 
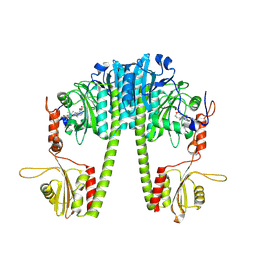 | | Cryo-EM structure of the full-length Pseudomonas aeruginosa bacteriophytochrome in its Pfr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Bodizs, S, Westenhoff, S. | | Deposit date: | 2024-03-28 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structures of a bathy phytochrome histidine kinase reveal a unique light-dependent activation mechanism.
Structure, 2024
|
|
9EUT
 
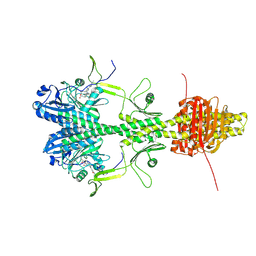 | | Cryo-EM structure of the full-length Pseudomonas aeruginosa bacteriophytochrome in its Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Bodizs, S, Westenhoff, S. | | Deposit date: | 2024-03-28 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structures of a bathy phytochrome histidine kinase reveal a unique light-dependent activation mechanism.
Structure, 2024
|
|
8ZM2
 
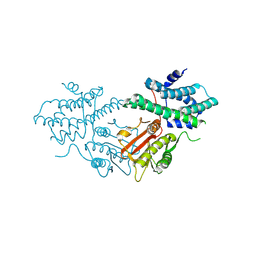 | | Structure of human pyruvate dehydrogenase kinase 2 complexed with compound 16 | | Descriptor: | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial, methyl (9~{R})-9-oxidanyl-9-(trifluoromethyl)fluorene-4-carboxylate | | Authors: | Akai, S, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-22 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Design and synthesis of novel fluorene derivatives as inhibitors of pyruvate dehydrogenase kinase.
Bioorg.Med.Chem.Lett., 109, 2024
|
|
8ZM1
 
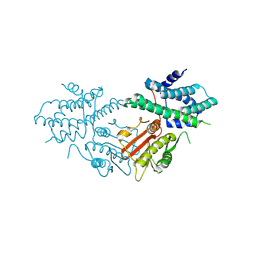 | | Structure of human pyruvate dehydrogenase kinase 2 complexed with compound 6 | | Descriptor: | (5~{R})-5-propan-2-ylindeno[1,2-b]pyridin-5-ol, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial | | Authors: | Akai, S, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-22 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Design and synthesis of novel fluorene derivatives as inhibitors of pyruvate dehydrogenase kinase.
Bioorg.Med.Chem.Lett., 109, 2024
|
|
8YON
 
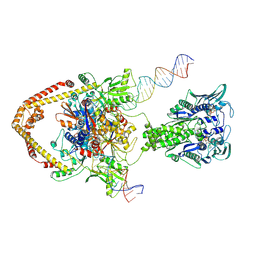 | |
8YOD
 
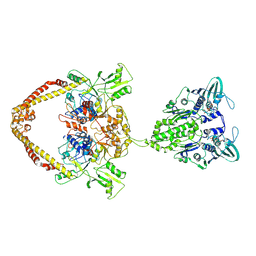 | |
8YO7
 
 | |
8YO5
 
 | |
8YO1
 
 | |
8YLU
 
 | | structure of phage T6 topoisomerase II central domain bound with DNA | | Descriptor: | DNA (5'-D(P*TP*AP*TP*AP*TP*GP*TP*GP*TP*AP*TP*AP*TP*AP*TP*AP*CP*AP*CP*AP*CP*A)-3'), DNA topoisomerase (ATP-hydrolyzing), DNA topoisomerase medium subunit, ... | | Authors: | Chen, Y.T, Xin, Y.H. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | structure of phage T6 topoisomerase II central domain bound with DNA
To be published
|
|
8YB4
 
 | | Pfr conformer of Arabidopsis thaliana phytochrome B in complex with phytochrome-interacting factor 6 | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, phytochrome B, phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
8UQK
 
 | |
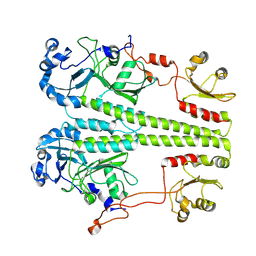
8UQI
 
 | |
8UPM
 
 | |
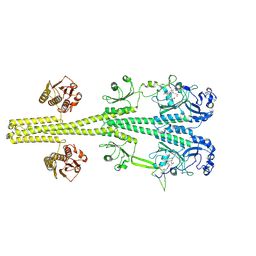
8UPK
 
 | |
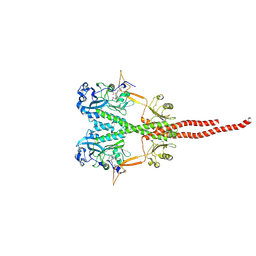
8UPH
 
 | |
8U8Z
 
 | | Cryo-EM structure of PsBphP in Pr state, extended DHp | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-18 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U65
 
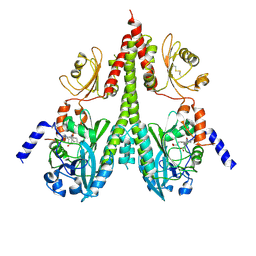 | | Cryo-EM structure of PsBphP in Pfr state, splayed PSM only | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U64
 
 | | Cryo-EM structure of PsBphP in Pfr state, medial PSM only | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U63
 
 | | Cryo-EM structure of PsBphP in Pfr state, Dimer of Dimers PSM only | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U62
 
 | | Cryo-EM structure of PsBphP in Pfr state, Dimer of Dimers FL | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U4X
 
 | | Cryo-EM structure of PsBphP in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-11 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
