5ZWL
 
 | |
1FS0
 
 | |
6CP3
 
 | | Monomer yeast ATP synthase (F1Fo) reconstituted in nanodisc with inhibitor of oligomycin bound. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
3FKS
 
 | | Yeast F1 ATPase in the absence of bound nucleotides | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Kabaleeswaran, V, Symersky, J, Shen, H, Walker, J.E, Leslie, A.G.W, Mueller, D.M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.587 Å) | | Cite: | Asymmetric structure of the yeast f1 ATPase in the absence of bound nucleotides.
J.Biol.Chem., 284, 2009
|
|
5IK2
 
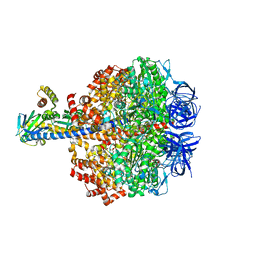 | | Caldalaklibacillus thermarum F1-ATPase (epsilon mutant) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Ferguson, S.A, Cook, G.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2016-03-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6TMK
 
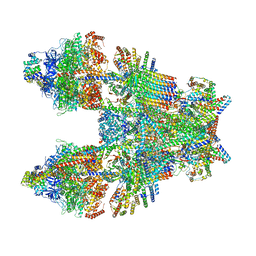 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, composite model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
1QO1
 
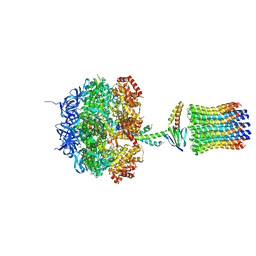 | | Molecular Architecture of the Rotary Motor in ATP Synthase from Yeast Mitochondria | | Descriptor: | ATP SYNTHASE ALPHA CHAIN, ATP SYNTHASE BETA CHAIN, ATP SYNTHASE DELTA CHAIN, ... | | Authors: | Stock, D, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1999-11-01 | | Release date: | 1999-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Molecular Architecture of the Rotary Motor in ATP Synthase
Science, 286, 1999
|
|
2W6G
 
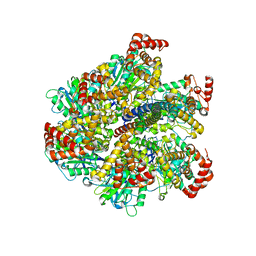 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 3. | | Descriptor: | ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT BETA, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Improving Diffraction by Humidity Control: A Novel Device Compatible with X-Ray Beamlines.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6TML
 
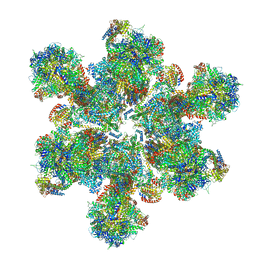 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase hexamer, composite model | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
2W6H
 
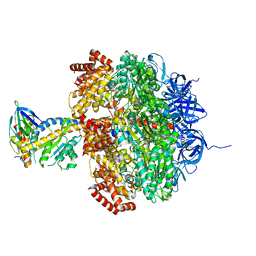 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 4A. | | Descriptor: | ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT BETA, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Improving diffraction by humidity control: a novel device compatible with X-ray beamlines.
Acta Crystallogr. D Biol. Crystallogr., 65, 2009
|
|
1H8H
 
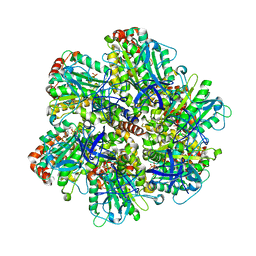 | | Bovine mitochondrial F1-ATPase crystallised in the presence of 5mm AMPPNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | Braig, K, Menz, R.I, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2001-02-06 | | Release date: | 2001-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure and Nucleotide Occupancy of Bovine Mitochondrial F(1)-ATPase are not Influenced by Crystallisation at High Concentrations of Nucleotide
FEBS Lett., 494, 2001
|
|
1OHH
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE complexed with the inhibitor protein IF1 | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Cabezon, E, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Bovine F1-ATPase in Complex with its Regulatory Protein If1
Nat.Struct.Biol., 10, 2003
|
|
1H8E
 
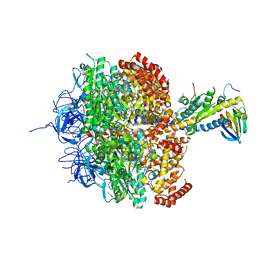 | | (ADP.AlF4)2(ADP.SO4) bovine F1-ATPase (all three catalytic sites occupied) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, GLYCEROL, ... | | Authors: | Menz, R.I, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2001-02-02 | | Release date: | 2001-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase with Nucleotide Bound to All Three Catalytic Sites: Implications for the Mechanism of Rotary Catalysis
Cell(Cambridge,Mass.), 106, 2001
|
|
1MAB
 
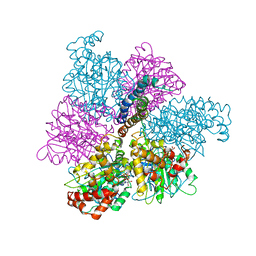 | | RAT LIVER F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bianchet, M.A, Amzel, L.M. | | Deposit date: | 1998-08-06 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8-A structure of rat liver F1-ATPase: configuration of a critical intermediate in ATP synthesis/hydrolysis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1W0K
 
 | | ADP inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, MITOCHONDRIAL PRECURSOR, ... | | Authors: | Kagawa, R, Montgomery, M.G, Braig, K, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Structure of Bovine F1-ATPase Inhibited by Adp and Beryllium Fluoride
Embo J., 23, 2004
|
|
1W0J
 
 | | Beryllium fluoride inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, MITOCHONDRIAL PRECURSOR, ... | | Authors: | Kagawa, R, Montgomery, M.G, Braig, K, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Bovine F1-ATPase Inhibited by Adp and Beryllium Fluoride
Embo J., 23, 2004
|
|
1NBM
 
 | | THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, F1-ATPASE, ... | | Authors: | Orriss, G.L, Leslie, A.G.W, Braig, K, Walker, J.E. | | Deposit date: | 1998-04-30 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism.
Structure, 6, 1998
|
|
4TT3
 
 | | The Pathway of Binding of the Intrinsically Disordered Mitochondrial Inhibitor Protein to F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2014-06-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TSF
 
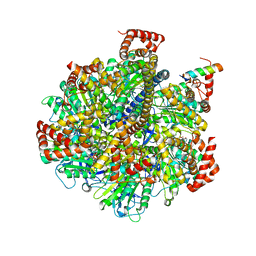 | | The Pathway of Binding of the Intrinsically Disordered Mitochondrial Inhibitor Protein to F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2QE7
 
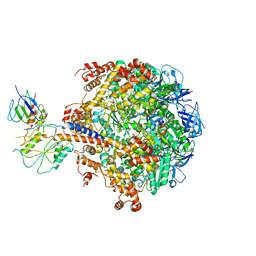 | | Crystal structure of the f1-atpase from the thermoalkaliphilic bacterium bacillus sp. ta2.a1 | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit epsilon, ... | | Authors: | Stocker, A, Keis, S, Vonck, J, Cook, G.M, Dimroth, P. | | Deposit date: | 2007-06-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Structural Basis for Unidirectional Rotation of Thermoalkaliphilic F(1)-ATPase.
Structure, 15, 2007
|
|
2V7Q
 
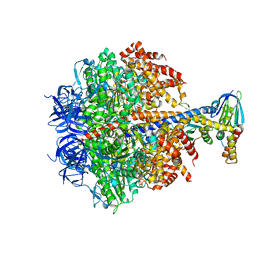 | | The structure of F1-ATPase inhibited by I1-60HIS, a monomeric form of the inhibitor protein, IF1. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP SYNTHASE DELTA CHAIN, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How the Regulatory Protein, If1, Inhibits F1- ATPase from Bovine Mitochondria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
5LQY
 
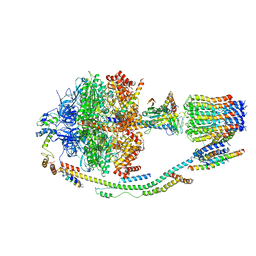 | | Structure of F-ATPase from Pichia angusta, in state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase OSCP subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LQX
 
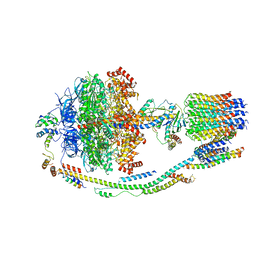 | | Structure of F-ATPase from Pichia angusta, state3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase AAP1 subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LQZ
 
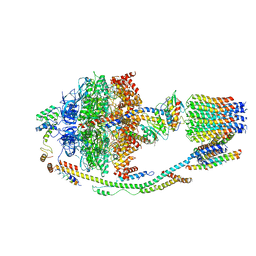 | | Structure of F-ATPase from Pichia angusta, state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase OSCP subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4XD7
 
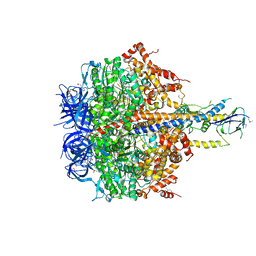 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
