6CTE
 
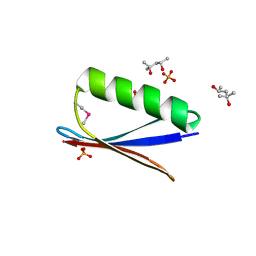 | | 77Se-NMR probes the protein environment of selenomethionine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Chen, Q, Rozovsky, S. | | Deposit date: | 2018-03-22 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6CQ6
 
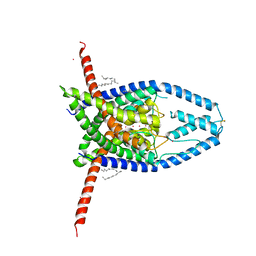 | | K2P2.1(TREK-1) apo structure | | Descriptor: | CADMIUM ION, DECANE, POTASSIUM ION, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2018-03-14 | | Release date: | 2018-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | K2P2.1 (TREK-1)-activator complexes reveal a cryptic selectivity filter binding site.
Nature, 547, 2017
|
|
7MZI
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 | | Descriptor: | GLYCEROL, Spike protein S1, TETRAETHYLENE GLYCOL, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
6H49
 
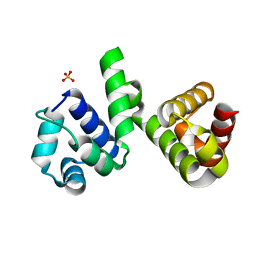 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | Descriptor: | Orf20, SULFATE ION | | Authors: | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
6CQG
 
 | |
8A9P
 
 | | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment | | Descriptor: | (3-phenyl-1,2,4-oxadiazol-5-yl)methanamine, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment
To Be Published
|
|
6H4D
 
 | | Crystal structure of RsgA from Pseudomonas aeruginosa | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Small ribosomal subunit biogenesis GTPase RsgA, ZINC ION | | Authors: | Rocchio, S, Santorelli, D, Travaglini-Allocatelli, C, Federici, L, Di Matteo, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional investigation of the Small Ribosomal Subunit Biogenesis GTPase A (RsgA) from Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6CQN
 
 | | Crystal structure of F5 TCR -DR11-RQ13 peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, F5 alpha chain, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-15 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
8A6W
 
 | |
6H2X
 
 | | MukB coiled-coil elbow from E. coli | | Descriptor: | Chromosome partition protein MukB,Chromosome partition protein MukB | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A folded conformation of MukBEF and cohesin.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6CR0
 
 | | 1.55 A resolution structure of (S)-6-hydroxynicotine oxidase from Shinella HZN7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-6-hydroxynicotine oxidase, ACETATE ION, ... | | Authors: | Deay III, D, Lovell, S, Battaile, K.P, Petillo, P, Richter, M. | | Deposit date: | 2018-03-16 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Improving the kinetic parameters of nicotine oxidizing enzymes by homologous structure comparison and rational design
Arch.Biochem.Biophys., 2022
|
|
7MZN
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 231 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 231 heavy chain, PDI 231 light chain, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7B28
 
 | |
7NLI
 
 | |
7R1I
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7B4Q
 
 | | Structure of a cold active HSL family esterase reveals mechanisms of low temperature adaptation and substrate specificity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Jones, D.D, Noby, N, Auhim, H. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure and in silico simulations of a cold-active esterase reveals its prime cold-adaptation mechanism.
Open Biology, 11, 2021
|
|
6H4Q
 
 | | Crystal structure of human KDM4A in complex with compound 34a | | Descriptor: | 8-[4-(1-methylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6CRN
 
 | | Structure of the USP15 deubiquitinase domain in complex with a high-affinity first-generation Ubv | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 15, Ubiquitin variant 15.2, ZINC ION | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-03-19 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
7AXQ
 
 | |
7R5B
 
 | | Crystal structure of BRD4(1) in complex with the inhibitor MPM2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1-(3-aminophenyl)-3-methyl-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrol-4-one, Bromodomain-containing protein 4, ... | | Authors: | Huegle, M. | | Deposit date: | 2022-02-10 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A novel pan-selective bromodomain inhibitor for epigenetic drug design
Eur.J.Med.Chem., 249, 2023
|
|
7NLH
 
 | |
6CS8
 
 | | High resolution crystal structure of FtsY-NG domain of E. coli | | Descriptor: | 1H-indole-6-carbonitrile, SODIUM ION, Signal recognition particle receptor FtsY | | Authors: | Faoro, C, Ataide, S.F. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Discovery of fragments that target key interactions in the signal recognition particle (SRP) as potential leads for a new class of antibiotics.
PLoS ONE, 13, 2018
|
|
6H5W
 
 | |
6H52
 
 | | Crystal structure of human KDM5B in complex with compound 34g | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-(1-cyclopentylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6CEA
 
 | | Crystal structure of fragment 3-(quinolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(quinolin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
