1V0O
 
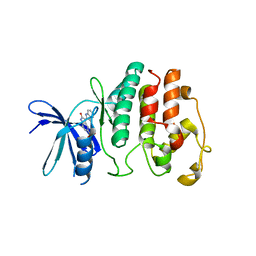 | | Structure of P. falciparum PfPK5-Indirubin-5-sulphonate ligand complex | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, CELL DIVISION CONTROL PROTEIN 2 HOMOLOG | | Authors: | Holton, S, Merckx, A, Burgess, D, Doerig, C, Noble, M, Endicott, J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of P. Falciparum Pfpk5 Test the Cdk Regulation Paradigm and Suggest Mechanisms of Small Molecule Inhibition
Structure, 11, 2003
|
|
1V0P
 
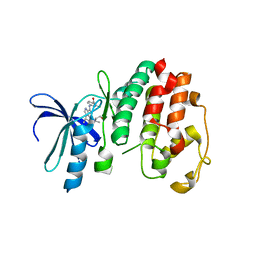 | | Structure of P. falciparum PfPK5-Purvalanol B ligand complex | | Descriptor: | CELL DIVISION CONTROL PROTEIN 2 HOMOLOG, PURVALANOL B | | Authors: | Holton, S, Merckx, A, Burgess, D, Doerig, C, Noble, M, Endicott, J. | | Deposit date: | 2004-04-01 | | Release date: | 2004-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of P. Falciparum Pfpk5 Test the Cdk Regulation Paradigm and Suggest Mechanisms of Small Molecule Inhibition
Structure, 11, 2003
|
|
1V0R
 
 | |
1V0S
 
 | |
1V0T
 
 | |
1V0U
 
 | |
1V0V
 
 | |
1V0W
 
 | |
1V0Y
 
 | |
1V0Z
 
 | | Structure of Neuraminidase from English duck subtype N6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Rudino-Pinera, E, Tunnah, P, Crennell, S.J, Webster, R.G, Laver, W.G, Garman, E.F. | | Deposit date: | 2004-03-12 | | Release date: | 2006-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Crystal Structure of Influenza Type a Virus Neuraminidase of the N6 Subtype at 1.85 A Resolution
To be Published
|
|
1V10
 
 | | Structure of Rigidoporus lignosus laccase from hemihedrally twinned crystals | | Descriptor: | COPPER (II) ION, LACCASE | | Authors: | Rizzi, M, Garavaglia, S, Palmieri, F, Cambria, A. | | Deposit date: | 2004-04-02 | | Release date: | 2004-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Rigidoporus Lignosus Laccase Containing a Full Complement of Copper Ions, Reveals an Asymmetrical Arrangement for the T3 Copper Pair
J.Mol.Biol., 342, 2004
|
|
1V11
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
1V13
 
 | |
1V14
 
 | |
1V15
 
 | |
1V16
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-07 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
1V18
 
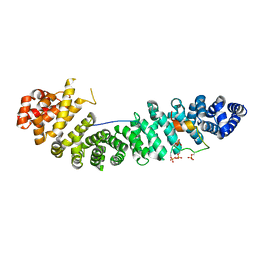 | |
1V19
 
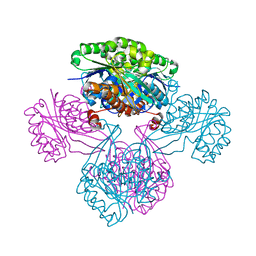 | | 2-KETO-3-DEOXYGLUCONATE KINASE FROM THERMUS THERMOPHILUS | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-KETO-3-DEOXYGLUCONATE KINASE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2004-04-12 | | Release date: | 2004-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Thermus Thermophilus 2-Keto-3-Deoxygluconate Kinase: Evidence for Recognition of an Open Chain Substrate
J.Mol.Biol., 340, 2004
|
|
1V1A
 
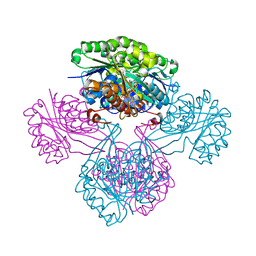 | |
1V1B
 
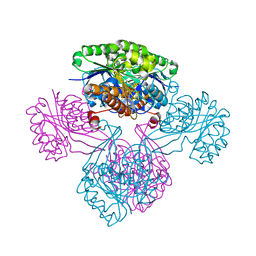 | |
1V1C
 
 | |
1V1D
 
 | |
1V1F
 
 | | Structure of the Arabidopsis thaliana SOS3 complexed with Calcium(II) and Manganese(II) ions | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCINEURIN B-LIKE PROTEIN 4, CALCIUM ION, ... | | Authors: | Sanchez-Barrena, M.J, Martinez-Ripoll, M, Zhu, J.K, Albert, A. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of the Arabidopsis Thaliana SOS3: Molecular Mechanism of Sensing Calcium for Salt Stress Response
J.Mol.Biol., 345, 2005
|
|
1V1G
 
 | | Structure of the Arabidopsis thaliana SOS3 complexed with Calcium(II) ion | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCINEURIN B-LIKE PROTEIN 4, CALCIUM ION, ... | | Authors: | Sanchez-Barrena, M.J, Martinez-Ripoll, M, Zhu, J.K, Albert, A. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of the Arabidopsis Thaliana SOS3: Molecular Mechanism of Sensing Calcium for Salt Stress Response
J.Mol.Biol., 345, 2005
|
|
1V1H
 
 | | Adenovirus fibre shaft sequence N-terminally fused to the bacteriophage T4 fibritin foldon trimerisation motif with a short linker | | Descriptor: | FIBRITIN, FIBER PROTEIN | | Authors: | Papanikolopoulou, K, Teixeira, S, Belrhali, H, Forsyth, V.T, Mitraki, A, van Raaij, M.J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-07-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenovirus Fibre Shaft Sequences Fold Into the Native Triple Beta-Spiral Fold When N-Terminally Fused to the Bacteriophage T4 Fibritin Foldon Trimerisation Motif
J.Mol.Biol., 342, 2004
|
|
