4IAT
 
 | |
3NUZ
 
 | |
3NWP
 
 | |
4IF4
 
 | |
4IKI
 
 | |
4IKJ
 
 | |
4GN4
 
 | |
4GND
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains | | Descriptor: | Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
3MYX
 
 | |
4GT6
 
 | |
4GPK
 
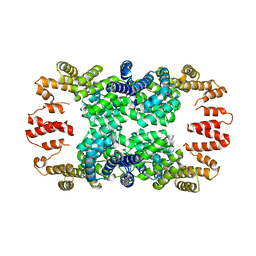 | | Crystal structure of NprR in complex with its cognate peptide NprX | | Descriptor: | NprR, NprX peptide | | Authors: | Zouhir, S, Guimaraes, B, Perchat, S, Nicaise, M, Lereclus, D, Nessler, S. | | Deposit date: | 2012-08-21 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR.
Nucleic Acids Res., 41, 2013
|
|
3N0W
 
 | |
3MX3
 
 | |
3MTQ
 
 | |
3N0X
 
 | |
5K2H
 
 | |
3NNB
 
 | |
3NRE
 
 | |
4GZV
 
 | |
4H1U
 
 | | Nucleotide-free human dynamin-1-like protein GTPase-GED fusion | | Descriptor: | CITRATE ANION, Dynamin-1-like protein | | Authors: | Wenger, J, Klinglmayr, E, Puehringer, S, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
5K2F
 
 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4H1V
 
 | | GMP-PNP bound dynamin-1-like protein GTPase-GED fusion | | Descriptor: | Dynamin-1-like protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Wenger, J, Klinglmayr, E, Eibl, C, Hessenberger, M, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
4H6I
 
 | |
4IZS
 
 | |
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
