1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1G7H
 
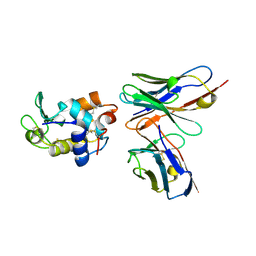 | | CRYSTAL STRUCTURE OF HEN EGG WHITE LYSOZYME (HEL) COMPLEXED WITH THE MUTANT ANTI-HEL MONOCLONAL ANTIBODY D1.3(VLW92A) | | Descriptor: | ANTI-HEN EGG WHITE LYSOZYME MONOCLONAL ANTIBODY D1.3, LYSOZYME C | | Authors: | Sundberg, E.J, Urrutia, M, Braden, B.C, Isern, J, Mariuzza, R.A. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Estimation of the hydrophobic effect in an antigen-antibody protein-protein interface.
Biochemistry, 39, 2000
|
|
1G7J
 
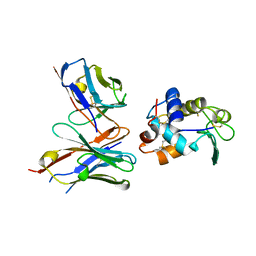 | | CRYSTAL STRUCTURE OF HEN EGG WHITE LYSOZYME (HEL) COMPLEXED WITH THE MUTANT ANTI-HEL MONOCLONAL ANTIBODY D1.3 (VLW92H) | | Descriptor: | ANTI-HEN EGG WHITE LYSOZYME MONOCLONAL ANTIBODY D1.3, LYSOZYME C | | Authors: | Sundberg, E.J, Urrutia, M, Braden, B.C, Isern, J, Mariuzza, R.A. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Estimation of the hydrophobic effect in an antigen-antibody protein-protein interface.
Biochemistry, 39, 2000
|
|
1K5K
 
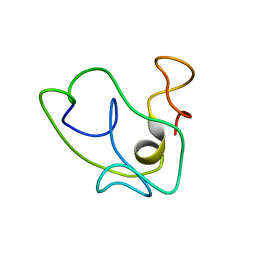 | | Homonuclear 1H Nuclear Magnetic Resonance Assignment and Structural Characterization of HIV-1 Tat Mal Protein | | Descriptor: | TAT protein | | Authors: | Gregoire, C, Peloponese, J.M, Esquieu, D, Opi, S, Campbell, G, Solomiac, M, Lebrun, E, Lebreton, J, Loret, E.P. | | Deposit date: | 2001-10-11 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Homonuclear (1)H-NMR assignment and structural characterization of human immunodeficiency virus type 1 Tat Mal protein.
Biopolymers, 62, 2001
|
|
1U4G
 
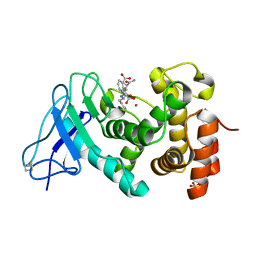 | | Elastase of Pseudomonas aeruginosa with an inhibitor | | Descriptor: | CALCIUM ION, Elastase, N-(1-CARBOXY-3-PHENYLPROPYL)PHENYLALANYL-ALPHA-ASPARAGINE, ... | | Authors: | Bitto, E, McKay, D.B. | | Deposit date: | 2004-07-25 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Elastase of Pseudomonas aeruginosa with an inhibitor
To be Published, 2004
|
|
1K43
 
 | | 10 Structure Ensemble of the 14-residue peptide RG-KWTY-NG-ITYE-GR (MBH12) | | Descriptor: | MBH12 | | Authors: | Pastor, M.T, Lopez de la Paz, M, Lacroix, E, Serrano, L, Perez-Paya, E. | | Deposit date: | 2001-10-05 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Combinatorial approaches: a new tool to search for highly structured beta-hairpin peptides.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4H90
 
 | | Radiation damage study of lysozyme - 0.28 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1999 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1K3O
 
 | | Crystal Structure Analysis of apo Glutathione S-Transferase | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
6HEB
 
 | | Influenza A Virus N9 Neuraminidase complex with Oseltamivir (Tern). | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-20 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site.
To Be Published
|
|
4EOL
 
 | | Thr 160 phosphorylated CDK2 H84S, Q85M, K89D - human cyclin A3 complex with the inhibitor RO3306 | | Descriptor: | (5E)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOQ
 
 | | Thr 160 phosphorylated CDK2 WT - human cyclin A3 complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
1K6J
 
 | | Crystal structure of Nmra, a negative transcriptional regulator (Monoclinic form) | | Descriptor: | CHLORIDE ION, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2001
|
|
3I50
 
 | | Crystal structure of the West Nile Virus envelope glycoprotein in complex with the E53 antibody Fab | | Descriptor: | Envelope glycoprotein, murine heavy chain (IgG3) of E53 monoclonal antibody Fab, murine kappa light chain of E53 monoclonal antibody Fab | | Authors: | Nybakken, G.E, Warren, J.T, Chen, B.R, Nelson, C.A, Fremont, D.H. | | Deposit date: | 2009-07-03 | | Release date: | 2009-10-27 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody.
Embo J., 28, 2009
|
|
2OFC
 
 | | The crystal structure of Sclerotium rolfsii lectin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural Basis for the Carbohydrate Recognition of the Sclerotium rolfsii Lectin
J.Mol.Biol., 368, 2007
|
|
6HFC
 
 | | Influenza A Virus N9 Neuraminidase Native (Tern). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
2G5T
 
 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) complexed with cyanopyrrolidine (C5-pro-pro) inhibitor 21ag | | Descriptor: | 3-{[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHOXY}-4-CHLOROBENZOIC ACID, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Fry, E.H, Lake, M.R, Solomon, L.R, Pei, Z, Li, X. | | Deposit date: | 2006-02-23 | | Release date: | 2006-07-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery, structure-activity relationship, and pharmacological evaluation of (5-substituted-pyrrolidinyl-2-carbonyl)-2-cyanopyrrolidines as potent dipeptidyl peptidase IV inhibitors.
J.Med.Chem., 49, 2006
|
|
3MXF
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective inhibition of BET bromodomains.
Nature, 468, 2010
|
|
1K8J
 
 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
4MRY
 
 | | Crystal Structure of Ca(2+)- discharged Y138F obelin mutant from Obelia longissima at 1.30 Angstrom resolution | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Natashin, P.V, Ding, W, Eremeeva, E.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Structures of the Ca2+-regulated photoprotein obelin Y138F mutant before and after bioluminescence support the catalytic function of a water molecule in the reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6G0A
 
 | |
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
4H92
 
 | | Radiation damage study of lysozyme- 0.42 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2002 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3HNJ
 
 | | Crystal structure of the Zn-induced tetramer of the engineered cyt cb562 variant RIDC-2 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ZINC ION | | Authors: | Salgado, E.N, Lewis, R.A, Brodin, J, Tezcan, F.A. | | Deposit date: | 2009-05-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal templated design of protein interfaces.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6HHP
 
 | | Ternary complex of Estrogen Receptor alpha peptide and 14-3-3 sigma C42 mutant bound to disulfide fragment PPI stabilizer 1 | | Descriptor: | (1~{R})-2-(4-chloranylphenoxy)-2-methyl-1-[methyl(2-sulfanylethyl)amino]propan-1-ol, 14-3-3 protein sigma, Estrogen receptor, ... | | Authors: | Sijbesma, E, Hallenbeck, K.K, Leysen, S, Arkin, M.R, Ottmann, C. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Site-Directed Fragment-Based Screening for the Discovery of Protein-Protein Interaction Stabilizers.
J. Am. Chem. Soc., 141, 2019
|
|
6HON
 
 | | Drosophila NOT4 CBM peptide bound to human CAF40 | | Descriptor: | CCR4-NOT transcription complex subunit 4, isoform L, CCR4-NOT transcription complex subunit 9, ... | | Authors: | Raisch, T, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A conserved CAF40-binding motif in metazoan NOT4 mediates association with the CCR4-NOT complex.
Genes Dev., 33, 2019
|
|
