5CIK
 
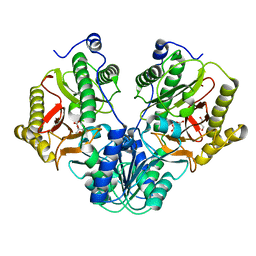 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris in citrate condition | | Descriptor: | CITRIC ACID, GLYCEROL, Proline dipeptidase | | Authors: | Kumar, A, Are, V, Ghosh, B, Agrawal, U, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris in citrate condition
To Be Published
|
|
3CV3
 
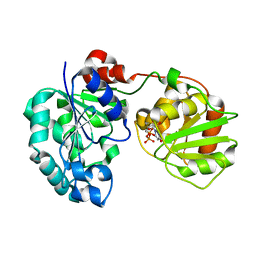 | | Crystal Structure of GumK mutant D157A in complex with UDP | | Descriptor: | Glucuronosyltransferase GumK, URIDINE-5'-DIPHOSPHATE | | Authors: | Barreras, M. | | Deposit date: | 2008-04-17 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and mechanism of GumK, a membrane-associated glucuronosyltransferase.
J.Biol.Chem., 283, 2008
|
|
3CUY
 
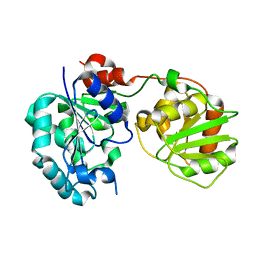 | | Crystal Structure of GumK mutant D157A | | Descriptor: | Glucuronosyltransferase GumK | | Authors: | Barreras, M. | | Deposit date: | 2008-04-17 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of GumK, a membrane-associated glucuronosyltransferase.
J.Biol.Chem., 283, 2008
|
|
2E12
 
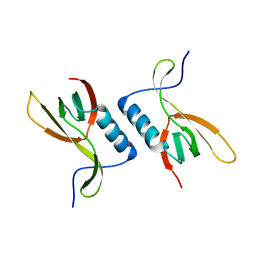 | | The crystal structure of XC5848 from Xanthomonas campestris adopting a novel variant of Sm-like motif | | Descriptor: | Hypothetical protein XCC3642 | | Authors: | Chin, K.-H, Ruan, S.-K, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2006-10-17 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | XC5848, an ORFan protein from Xanthomonas campestris, adopts a novel variant of Sm-like motif
Proteins, 68, 2007
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
8B06
 
 | | TRYPTOPHAN SYNTHASE - Cryo-trapping by the spitrobot crystal plunger after 25 sec | | Descriptor: | CESIUM ION, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Dopke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-09-07 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8B03
 
 | | TRYPTOPHAN SYNTHASE - Cryo-trapping by the spitrobot crystal plunger after 0 sec | | Descriptor: | CESIUM ION, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Dopke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-09-07 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8B08
 
 | | TRYPTOPHAN SYNTHASE - Cryo-trapping by the spitrobot crystal plunger after 30 sec | | Descriptor: | CESIUM ION, CHLORIDE ION, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Dopke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-09-07 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8B05
 
 | | TRYPTOPHAN SYNTHASE - Cryo-trapping by the spitrobot crystal plunger after 20 sec | | Descriptor: | CESIUM ION, CHLORIDE ION, INDOLE, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Dopke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-09-07 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWC
 
 | | Xylose Isomerase in 85% relative humidity environment | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AW9
 
 | | Xylose Isomerase in 75% relative humidity environment | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AW8
 
 | | Xylose Isomerase in 70% relative humidity environment | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWE
 
 | | Xylose Isomerase in 99% relative humidity environment | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWD
 
 | | Xylose Isomerase in 95% relative humidity environment | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWF
 
 | | Xylose Isomerase in 80% relative humidity environment | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWU
 
 | | Millisecond cryo-trapping by the spitrobot crystal plunger, Xylose Isomerase with Glucose at 250ms | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWS
 
 | | Millisecond cryo-trapping by the spitrobot crystal plunger, Xylose Isomerase with Glucose at 50ms | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWV
 
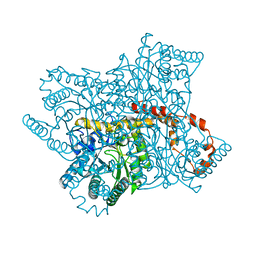 | | Millisecond cryo-trapping by the spitrobot crystal plunger, Xylose Isomerase with Glucose at 500ms | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWX
 
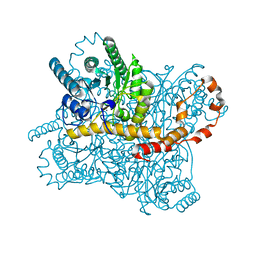 | | Millisecond cryo-trapping by the spitrobot crystal plunger, Xylose Isomerase with Glucose at 1s | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWY
 
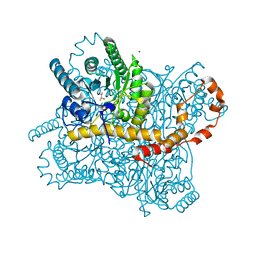 | | Millisecond cryo-trapping by the spitrobot crystal plunger, Serial measurement Xylose Isomerase with 2,3-butanediol at 50ms | | Descriptor: | MAGNESIUM ION, Meso-2,3-Butanediol, Xylose isomerase | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8B3M
 
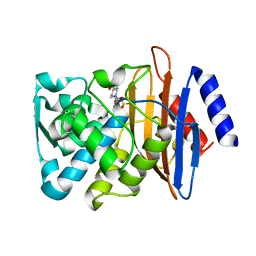 | | Millisecond cryo-trapping by the spitrobot crystal plunger, CTXM-14 Avibactam complex, SSX, 1 sec | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
8AWB
 
 | | Xylose Isomerase in 90% relative humidity environment | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
6XCT
 
 | | Porcine pepsin in complex with amprenavir | | Descriptor: | Pepsin A, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Porcine pepsin in complex with amprenavir
To Be Published
|
|
7QFO
 
 | | Human Topoisomerase II Beta ATPase AMPPNP | | Descriptor: | ALANINE, DNA topoisomerase 2-beta, MAGNESIUM ION, ... | | Authors: | Ling, E.M, Basle, A, Cowell, I.G, Blower, T.R, Austin, C.A. | | Deposit date: | 2021-12-06 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A comprehensive structural analysis of the ATPase domain of human DNA topoisomerase II beta bound to AMPPNP, ADP, and the bisdioxopiperazine, ICRF193.
Structure, 30, 2022
|
|
1K19
 
 | | NMR Solution Structure of the Chemosensory Protein CSP2 from Moth Mamestra brassicae | | Descriptor: | Chemosensory Protein CSP2 | | Authors: | Mosbah, A, Campanacci, V, Lartigue, A, Tegoni, M, Cambillau, C, Darbon, H. | | Deposit date: | 2001-09-24 | | Release date: | 2002-12-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a chemosensory protein from the moth Mamestra brassicae
BIOCHEM.J., 369, 2003
|
|
