1UXE
 
 | | ADENOVIRUS AD37 FIBRE HEAD | | Descriptor: | ACETATE ION, FIBER PROTEIN, ZINC ION | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
1UUZ
 
 | | IVY:A NEW FAMILY OF PROTEIN | | Descriptor: | INHIBITOR OF VERTEBRATE LYSOZYME, LYSOZYME C | | Authors: | Abergel, C, Lembo, F, Byrne, D, Maza, C, Claverie, J.M. | | Deposit date: | 2004-01-12 | | Release date: | 2004-01-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Evolution of the Ivy Protein Family, Unexpected Lysozyme Inhibitors in Gram-Negative Bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1UOB
 
 | | Deacetoxycephalosporin C synthase complexed with 2-oxoglutarate and penicillin G | | Descriptor: | 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, ... | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UVB
 
 | | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza sativa | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Cheng, H.-C, Cheng, P.-T, Peng, P, Lyu, P.-C, Sun, Y.-J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza Sativa
Protein Sci., 13, 2004
|
|
5T78
 
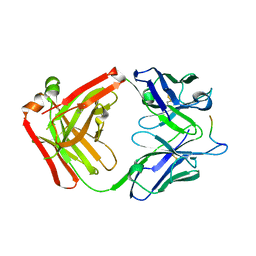 | | Crystal structure of therapeutic mAB AR20.5 in complex with MUC1 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Fab Fragment - AR20.5 - Heavy chain, Fab fragment AR20.5 - Light Chain, ... | | Authors: | Brooks, C.L, Movahedin, M. | | Deposit date: | 2016-09-02 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glycosylation of MUC1 influences the binding of a therapeutic antibody by altering the conformational equilibrium of the antigen.
Glycobiology, 27, 2017
|
|
1UUW
 
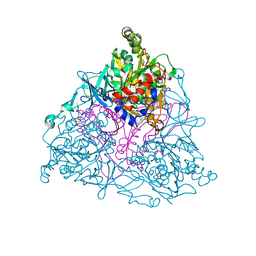 | | NAPHTHALENE 1,2-DIOXYGENASE WITH NITRIC OXIDE BOUND IN THE ACTIVE SITE. | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, NAPHTHALENE 1,2-DIOXYGENASE ALPHA SUBUNIT, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2004-01-11 | | Release date: | 2005-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | No Binding to Naphthalene Dioxygenase.
J.Biol.Inorg.Chem., 10, 2005
|
|
1UNB
 
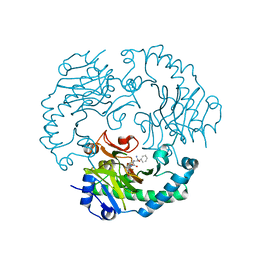 | | Deacetoxycephalosporin C synthase complexed with 2-oxoglutarate and ampicillin | | Descriptor: | (2S,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, ... | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-09 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UOG
 
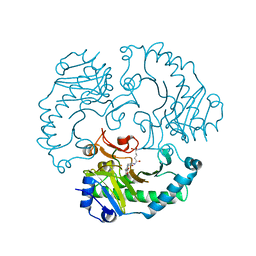 | | Deacetoxycephalosporin C synthase complexed with deacetoxycephalosporin C | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHETASE, DEACETOXYCEPHALOSPORIN-C, FE (II) ION | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-17 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
5T9A
 
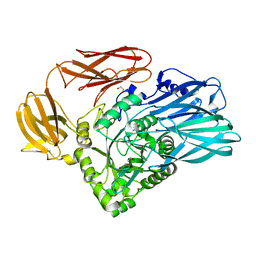 | | Crystal structure of BuGH2Cwt | | Descriptor: | 1,2-ETHANEDIOL, Glycoside Hydrolase, SULFATE ION | | Authors: | Pluvinage, B, Boraston, A.B, Abbott, W.D. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
1UKE
 
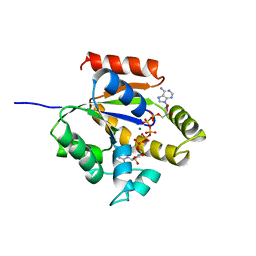 | | UMP/CMP KINASE FROM SLIME MOLD | | Descriptor: | MAGNESIUM ION, P1-(ADENOSINE-5'-P5-(URIDINE-5')PENTAPHOSPHATE, URIDYLMONOPHOSPHATE/CYTIDYLMONOPHOSPHATE KINASE | | Authors: | Scheffzek, K, Kliche, W, Wiesmueller, L, Reinstein, J. | | Deposit date: | 1998-01-07 | | Release date: | 1998-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the complex of UMP/CMP kinase from Dictyostelium discoideum and the bisubstrate inhibitor P1-(5'-adenosyl) P5-(5'-uridyl) pentaphosphate (UP5A) and Mg2+ at 2.2 A: implications for water-mediated specificity.
Biochemistry, 35, 1996
|
|
1UUF
 
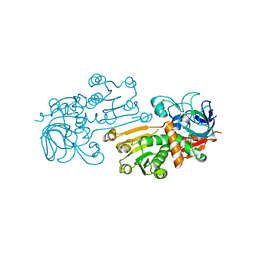 | |
1UPC
 
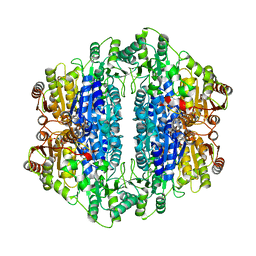 | | Carboxyethylarginine synthase from Streptomyces clavuligerus | | Descriptor: | CARBOXYETHYLARGININE SYNTHASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Caines, M.E.C, Elkins, J.M, Hewitson, K.S, Schofield, C.J. | | Deposit date: | 2003-09-29 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure and Mechanistic Implications of N2-(2-Carboxyethyl)Arginine Synthase, the First Enzyme in the Clavulanic Acid Biosynthesis Pathway
J.Biol.Chem., 279, 2004
|
|
5TBY
 
 | | HUMAN BETA CARDIAC HEAVY MEROMYOSIN INTERACTING-HEADS MOTIF OBTAINED BY HOMOLOGY MODELING (USING SWISS-MODEL) OF HUMAN SEQUENCE FROM APHONOPELMA HOMOLOGY MODEL (PDB-3JBH), RIGIDLY FITTED TO HUMAN BETA-CARDIAC NEGATIVELY STAINED THICK FILAMENT 3D-RECONSTRUCTION (EMD-2240) | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | ALAMO, L, WARE, J.S, PINTO, A, GILLILAN, R.E, SEIDMAN, J.G, SEIDMAN, C.E, PADRON, R. | | Deposit date: | 2016-09-13 | | Release date: | 2017-06-07 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes.
Elife, 6, 2017
|
|
5SY4
 
 | |
5SY9
 
 | |
1VPX
 
 | |
1W0R
 
 | |
1W0N
 
 | | Structure of uncomplexed Carbohydrate Binding Domain CBM36 | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, MAGNESIUM ION, ... | | Authors: | Jamal, S, Boraston, A.B, Davies, G.J. | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
5TCP
 
 | | Near-atomic resolution cryo-EM structure of the periplasmic domains of PrgH and PrgK | | Descriptor: | Lipoprotein PrgK, Protein PrgH | | Authors: | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
5SWJ
 
 | | Crystal Structure of ATPase delta1-79 Spa47 | | Descriptor: | Probable ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Burgess, J.L, Burgess, R.A, Dickenson, N.E. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation.
J. Biol. Chem., 291, 2016
|
|
5SYO
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with Cytisine | | Descriptor: | (1R,5S)-1,2,3,4,5,6-HEXAHYDRO-8H-1,5-METHANOPYRIDO[1,2-A][1,5]DIAZOCIN-8-ONE, Soluble acetylcholine receptor, Neuronal acetylcholine receptor subunit alpha-3 chimera | | Authors: | Bobango, J, Wu, J, Talley, I.T, Talley, T.T. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with Cytisine
To Be Published
|
|
5T9G
 
 | |
1W6P
 
 | | X-RAY CRYSTAL STRUCTURE OF C2S HUMAN GALECTIN-1 COMPLEXED WITH N- Acetyl-LACTOSAMINE | | Descriptor: | BETA-MERCAPTOETHANOL, GALECTIN-1, SULFATE ION, ... | | Authors: | Lopez-Lucendo, M.I.F, Gabius, H.J, Romero, A. | | Deposit date: | 2004-08-20 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Growth-Regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-Site Mutations and Their Impact on the Thermodynamics of Ligand Binding
J.Mol.Biol., 343, 2004
|
|
5T0V
 
 | | Architecture of the Yeast Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Sub-Complex Formed by the Iron Donor, Yfh1, and the Scaffold, Isu1 | | Descriptor: | Frataxin homolog, mitochondrial, Iron sulfur cluster assembly protein 1 | | Authors: | Ranatunga, W, Gakh, O, Galeano, B.K, Smith IV, D.Y, Soderberg, C.A, Al-Karadaghi, S, Thompson, J.R, Isaya, G. | | Deposit date: | 2016-08-16 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (17.5 Å) | | Cite: | Architecture of the Yeast Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Sub-Complex Formed by the Iron Donor, Yfh1, and the Scaffold, Isu1
J. Biol. Chem., 291, 2016
|
|
1VDD
 
 | |
